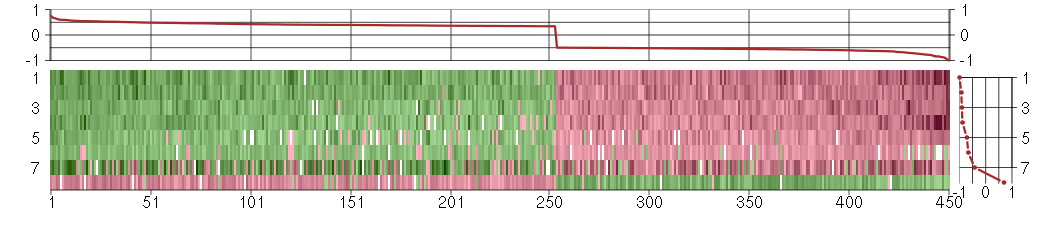

Under-expression is coded with green,
over-expression with red color.



ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.95 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: -0.7 ABRactive BCR-related gene (212895_s_at), score: 0.42 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: -0.6 ACP2acid phosphatase 2, lysosomal (202767_at), score: 0.4 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: -0.52 ACYP2acylphosphatase 2, muscle type (206833_s_at), score: 0.38 ADAadenosine deaminase (204639_at), score: 0.37 ADO2-aminoethanethiol (cysteamine) dioxygenase (212500_at), score: 0.4 AGPSalkylglycerone phosphate synthase (205401_at), score: -0.58 AHDC1AT hook, DNA binding motif, containing 1 (205002_at), score: 0.42 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: 0.4 AIM2absent in melanoma 2 (206513_at), score: -0.73 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.55 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.4 ANKHD1ankyrin repeat and KH domain containing 1 (208772_at), score: 0.36 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: -0.63 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: -0.56 APEX2APEX nuclease (apurinic/apyrimidinic endonuclease) 2 (204408_at), score: -0.5 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.35 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.55 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.5 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: 0.48 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.38 ARPP-19cyclic AMP phosphoprotein, 19 kD (214553_s_at), score: -0.56 ARSAarylsulfatase A (204443_at), score: 0.39 ASXL2additional sex combs like 2 (Drosophila) (218659_at), score: -0.55 ATP11BATPase, class VI, type 11B (212536_at), score: 0.4 ATP13A1ATPase type 13A1 (218052_s_at), score: 0.34 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.54 ATP6V1FATPase, H+ transporting, lysosomal 14kDa, V1 subunit F (201527_at), score: -0.51 ATP9AATPase, class II, type 9A (212062_at), score: 0.35 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: 0.45 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.62 BBXbobby sox homolog (Drosophila) (213016_at), score: -0.56 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.42 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: -0.5 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.77 BNIP1BCL2/adenovirus E1B 19kDa interacting protein 1 (204930_s_at), score: -0.5 BRD4bromodomain containing 4 (202102_s_at), score: 0.4 BTN2A2butyrophilin, subfamily 2, member A2 (205298_s_at), score: -0.54 BTRCbeta-transducin repeat containing (216091_s_at), score: -0.53 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.5 C11orf17chromosome 11 open reading frame 17 (219953_s_at), score: -0.56 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: -0.52 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.43 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.47 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.57 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.47 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.5 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.48 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.46 C6orf26chromosome 6 open reading frame 26 (212913_at), score: -0.5 CA3carbonic anhydrase III, muscle specific (204865_at), score: -0.63 CAMK2Gcalcium/calmodulin-dependent protein kinase II gamma (208095_s_at), score: 0.36 CANXcalnexin (208853_s_at), score: -0.5 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.35 CAPN1calpain 1, (mu/I) large subunit (200752_s_at), score: 0.35 CCNL2cyclin L2 (221427_s_at), score: -0.6 CD81CD81 molecule (200675_at), score: 0.37 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: -0.56 CDC2L2cell division cycle 2-like 2 (PITSLRE proteins) (212401_s_at), score: -0.54 CDCP1CUB domain containing protein 1 (218451_at), score: 0.34 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.36 CDKN2Acyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) (209644_x_at), score: 0.35 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.66 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: -0.6 CHPcalcium binding protein P22 (214665_s_at), score: 0.48 CHPFchondroitin polymerizing factor (202175_at), score: 0.4 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.4 CKBcreatine kinase, brain (200884_at), score: 0.34 CKLFchemokine-like factor (219161_s_at), score: -0.5 CLINT1clathrin interactor 1 (201768_s_at), score: -0.61 CLUAP1clusterin associated protein 1 (204577_s_at), score: -0.63 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (217947_at), score: -0.52 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (208912_s_at), score: 0.51 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.4 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.54 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.48 COX7A1cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) (204570_at), score: 0.38 CRATcarnitine acetyltransferase (209522_s_at), score: 0.4 CREBL2cAMP responsive element binding protein-like 2 (201990_s_at), score: 0.37 CRIP2cysteine-rich protein 2 (208978_at), score: 0.42 CRTAPcartilage associated protein (201380_at), score: 0.54 CRYABcrystallin, alpha B (209283_at), score: 0.47 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: 0.36 CST3cystatin C (201360_at), score: 0.4 CTBP1C-terminal binding protein 1 (213979_s_at), score: -0.61 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (201904_s_at), score: 0.36 CTNNBL1catenin, beta like 1 (221021_s_at), score: -0.56 CTTNcortactin (201059_at), score: 0.38 CUX1cut-like homeobox 1 (214743_at), score: 0.49 CXorf15chromosome X open reading frame 15 (219969_at), score: -0.54 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: 0.53 DAG1dystroglycan 1 (dystrophin-associated glycoprotein 1) (205417_s_at), score: 0.4 DAPdeath-associated protein (201095_at), score: 0.37 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.38 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.87 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.71 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: 0.52 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: 0.42 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (218579_s_at), score: -0.54 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: -0.51 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.61 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: -0.77 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.46 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: -0.52 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: 0.37 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: -0.54 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.4 E4F1E4F transcription factor 1 (218524_at), score: -0.5 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.41 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.36 EDNRAendothelin receptor type A (204464_s_at), score: 0.36 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.52 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: -0.56 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: -0.53 EPB41L3erythrocyte membrane protein band 4.1-like 3 (206710_s_at), score: 0.35 EPHB2EPH receptor B2 (209589_s_at), score: -0.56 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: 0.39 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: -0.61 EWSR1Ewing sarcoma breakpoint region 1 (209214_s_at), score: -0.58 FAF2Fas associated factor family member 2 (212106_at), score: -0.56 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.55 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.39 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: -0.52 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.75 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.43 FASNfatty acid synthase (212218_s_at), score: 0.38 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.5 FBN2fibrillin 2 (203184_at), score: 0.56 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.48 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.6 FOXG1forkhead box G1 (206018_at), score: -0.57 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: -0.62 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: -0.5 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.4 GAGE4G antigen 4 (207086_x_at), score: -0.9 GAGE6G antigen 6 (208155_x_at), score: -0.84 GATA3GATA binding protein 3 (209604_s_at), score: -0.78 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: -0.57 GHRgrowth hormone receptor (205498_at), score: 0.38 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: -0.59 GLSglutaminase (203159_at), score: 0.47 GLTPglycolipid transfer protein (219267_at), score: 0.52 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: 0.35 GNL3guanine nucleotide binding protein-like 3 (nucleolar) (217850_at), score: -0.51 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.42 GPR137BG protein-coupled receptor 137B (204137_at), score: 0.36 GSPT2G1 to S phase transition 2 (205541_s_at), score: 0.54 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.4 H1F0H1 histone family, member 0 (208886_at), score: -0.57 HAS2hyaluronan synthase 2 (206432_at), score: 0.48 HEPHhephaestin (203903_s_at), score: 0.35 HERC5hect domain and RLD 5 (219863_at), score: -0.74 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.55 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: -0.57 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.57 HIRIP3HIRA interacting protein 3 (204504_s_at), score: -0.53 HIST1H4Chistone cluster 1, H4c (205967_at), score: 0.55 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.39 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.46 HMGB1L10high-mobility group box 1-like 10 (216508_x_at), score: -0.5 HNRNPA1heterogeneous nuclear ribonucleoprotein A1 (214280_x_at), score: -0.54 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: -0.61 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: -0.64 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (200593_s_at), score: -0.55 HRASLSHRAS-like suppressor (219983_at), score: -0.53 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.56 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.4 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.34 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.54 IDEinsulin-degrading enzyme (203327_at), score: 0.38 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.6 IFI44interferon-induced protein 44 (214453_s_at), score: -0.52 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: -0.59 IFNAR2interferon (alpha, beta and omega) receptor 2 (204785_x_at), score: 0.35 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: 0.52 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: 0.36 IKIK cytokine, down-regulator of HLA II (200066_at), score: -0.56 IKZF5IKAROS family zinc finger 5 (Pegasus) (220086_at), score: -0.5 IL11interleukin 11 (206924_at), score: 0.37 IL21Rinterleukin 21 receptor (219971_at), score: 0.39 IMPDH1IMP (inosine monophosphate) dehydrogenase 1 (204169_at), score: 0.36 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.39 IPO8importin 8 (205701_at), score: -0.55 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.49 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.54 ITGBL1integrin, beta-like 1 (with EGF-like repeat domains) (214927_at), score: 0.34 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: 0.48 JMJD4jumonji domain containing 4 (218560_s_at), score: 0.34 KATNA1katanin p60 (ATPase-containing) subunit A 1 (205526_s_at), score: -0.51 KIAA0355KIAA0355 (203288_at), score: 0.36 KIAA0495KIAA0495 (213340_s_at), score: 0.68 KIAA1539KIAA1539 (207765_s_at), score: 0.38 KIF13Bkinesin family member 13B (202962_at), score: -0.55 KIF3Akinesin family member 3A (213623_at), score: -0.65 KIFC1kinesin family member C1 (209680_s_at), score: -0.58 KRIT1KRIT1, ankyrin repeat containing (34031_i_at), score: -0.57 KRT14keratin 14 (209351_at), score: 0.39 KRT19keratin 19 (201650_at), score: 0.47 KRT7keratin 7 (209016_s_at), score: 0.38 KTELC1KTEL (Lys-Tyr-Glu-Leu) containing 1 (218587_s_at), score: 0.4 LAMB3laminin, beta 3 (209270_at), score: -0.68 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.4 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.56 LIMK2LIM domain kinase 2 (202193_at), score: 0.41 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: 0.41 LMF1lipase maturation factor 1 (219135_s_at), score: 0.37 LOC151162hypothetical LOC151162 (212098_at), score: 0.45 LOC388796hypothetical LOC388796 (65588_at), score: -0.53 LOC727825hypothetical LOC727825 (220734_s_at), score: 0.35 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.52 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.51 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.59 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.4 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: 0.54 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.59 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.36 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.61 MACF1microtubule-actin crosslinking factor 1 (208633_s_at), score: -0.5 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.44 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: -0.57 MALLmal, T-cell differentiation protein-like (209373_at), score: 0.45 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.59 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: 0.49 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.56 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.72 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: -0.51 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: 0.4 MED6mediator complex subunit 6 (207079_s_at), score: -0.55 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.35 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.47 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.54 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.55 MLXIPMLX interacting protein (202519_at), score: 0.37 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: 0.37 MRC2mannose receptor, C type 2 (37408_at), score: 0.38 MREGmelanoregulin (219648_at), score: 0.42 MRFAP1L1Morf4 family associated protein 1-like 1 (212199_at), score: -0.54 MT1Gmetallothionein 1G (204745_x_at), score: 0.4 MT1Mmetallothionein 1M (217546_at), score: 0.46 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: 0.52 MT1Xmetallothionein 1X (204326_x_at), score: 0.45 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.36 MYO10myosin X (201976_s_at), score: 0.42 MYO1Cmyosin IC (214656_x_at), score: 0.75 MYO1Dmyosin ID (212338_at), score: 0.35 NACC2NACC family member 2, BEN and BTB (POZ) domain containing (212993_at), score: 0.4 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: -0.67 NBASneuroblastoma amplified sequence (202926_at), score: 0.44 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: -0.54 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.52 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: 0.38 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.78 NDNnecdin homolog (mouse) (209550_at), score: 0.44 NEFLneurofilament, light polypeptide (221805_at), score: -0.68 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.39 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (208003_s_at), score: -0.54 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.58 NKTRnatural killer-tumor recognition sequence (215338_s_at), score: -0.5 NMUneuromedin U (206023_at), score: -0.63 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: -0.5 NOL12nucleolar protein 12 (219324_at), score: -0.5 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.64 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.36 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.46 NR2F6nuclear receptor subfamily 2, group F, member 6 (209262_s_at), score: 0.39 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.63 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.51 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -1 ODF2outer dense fiber of sperm tails 2 (210415_s_at), score: -0.52 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.36 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.49 PARP4poly (ADP-ribose) polymerase family, member 4 (202239_at), score: -0.54 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.46 PCBP4poly(rC) binding protein 4 (209361_s_at), score: 0.41 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.35 PDAP1PDGFA associated protein 1 (202290_at), score: -0.52 PDE10Aphosphodiesterase 10A (205501_at), score: -0.5 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: 0.52 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: 0.4 PGK2phosphoglycerate kinase 2 (217009_at), score: -0.53 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.7 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.39 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.36 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.42 PNRC1proline-rich nuclear receptor coactivator 1 (209034_at), score: 0.34 PODXLpodocalyxin-like (201578_at), score: 0.41 PPCDCphosphopantothenoylcysteine decarboxylase (219066_at), score: -0.53 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: -0.52 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.63 PPP1R13Lprotein phosphatase 1, regulatory (inhibitor) subunit 13 like (218849_s_at), score: 0.35 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: 0.35 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: 0.35 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.37 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.47 PRKCHprotein kinase C, eta (218764_at), score: -0.5 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: -0.5 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.52 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: -0.51 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.34 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.48 PTNpleiotrophin (209466_x_at), score: -0.5 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.43 PXDNperoxidasin homolog (Drosophila) (212013_at), score: 0.51 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.39 R3HDM2R3H domain containing 2 (203831_at), score: 0.43 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.67 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.44 RBBP5retinoblastoma binding protein 5 (205169_at), score: -0.5 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: 0.46 RDXradixin (204969_s_at), score: -0.51 REEP1receptor accessory protein 1 (204364_s_at), score: -0.52 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.36 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: -0.52 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: 0.46 RHODras homolog gene family, member D (31846_at), score: 0.42 RIN2Ras and Rab interactor 2 (209684_at), score: 0.37 RNF19Aring finger protein 19A (220483_s_at), score: -0.58 RNPS1RNA binding protein S1, serine-rich domain (207939_x_at), score: -0.5 RPL15P22ribosomal protein L15 pseudogene 22 (217266_at), score: 0.36 RPL31ribosomal protein L31 (221593_s_at), score: 0.43 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: -0.52 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.51 RTN3reticulon 3 (219549_s_at), score: 0.36 SAFB2scaffold attachment factor B2 (32099_at), score: -0.57 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.43 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: -0.63 SC65synaptonemal complex protein SC65 (204078_at), score: 0.39 SCG5secretogranin V (7B2 protein) (203889_at), score: 0.46 SDC4syndecan 4 (202071_at), score: 0.39 SEC16ASEC16 homolog A (S. cerevisiae) (215696_s_at), score: 0.41 SEC61A1Sec61 alpha 1 subunit (S. cerevisiae) (217716_s_at), score: 0.39 SEC63SEC63 homolog (S. cerevisiae) (201914_s_at), score: 0.41 SENP2SUMO1/sentrin/SMT3 specific peptidase 2 (218122_s_at), score: -0.53 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.37 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.34 SF3B1splicing factor 3b, subunit 1, 155kDa (201070_x_at), score: -0.55 SF4splicing factor 4 (215004_s_at), score: -0.58 SFRS12IP1SFRS12-interacting protein 1 (217608_at), score: -0.53 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: -0.52 SFXN3sideroflexin 3 (220974_x_at), score: 0.37 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.46 SHFM1split hand/foot malformation (ectrodactyly) type 1 (202276_at), score: -0.59 SHMT1serine hydroxymethyltransferase 1 (soluble) (209980_s_at), score: -0.51 SIDT2SID1 transmembrane family, member 2 (56256_at), score: 0.42 SIRPAsignal-regulatory protein alpha (202897_at), score: 0.35 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.44 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: 0.59 SLC25A20solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 (203658_at), score: -0.52 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: 0.42 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.38 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.59 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: 0.36 SLC39A4solute carrier family 39 (zinc transporter), member 4 (219215_s_at), score: 0.35 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.47 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: 0.38 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.55 SMAD5SMAD family member 5 (205187_at), score: -0.57 SMAD7SMAD family member 7 (204790_at), score: 0.4 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.63 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.72 SNAPC1small nuclear RNA activating complex, polypeptide 1, 43kDa (205443_at), score: -0.53 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.81 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: 0.4 SPIN1spindlin 1 (217813_s_at), score: -0.51 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: 0.44 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: -0.53 SSNA1Sjogren syndrome nuclear autoantigen 1 (210378_s_at), score: -0.54 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.63 STOML2stomatin (EPB72)-like 2 (215416_s_at), score: -0.5 STX12syntaxin 12 (212112_s_at), score: 0.39 STX16syntaxin 16 (221499_s_at), score: -0.59 SUPT16Hsuppressor of Ty 16 homolog (S. cerevisiae) (217815_at), score: -0.52 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.59 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.45 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.58 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.51 TBC1D9BTBC1 domain family, member 9B (with GRAM domain) (215994_x_at), score: 0.38 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.75 TCFL5transcription factor-like 5 (basic helix-loop-helix) (204849_at), score: 0.4 TEAD1TEA domain family member 1 (SV40 transcriptional enhancer factor) (214600_at), score: -0.51 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.36 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: -0.58 TESK1testis-specific kinase 1 (204106_at), score: 0.42 TFGTRK-fused gene (217839_at), score: 0.36 TGIF1TGFB-induced factor homeobox 1 (203313_s_at), score: -0.5 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: -0.5 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.54 TMEM204transmembrane protein 204 (219315_s_at), score: 0.52 TMEM47transmembrane protein 47 (209656_s_at), score: 0.38 TMEM51transmembrane protein 51 (218815_s_at), score: -0.55 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.44 TMEM97transmembrane protein 97 (212279_at), score: -0.54 TMOD1tropomodulin 1 (203661_s_at), score: -0.67 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: 0.39 TNS1tensin 1 (221748_s_at), score: 0.51 TOB1transducer of ERBB2, 1 (202704_at), score: 0.53 TRIM16tripartite motif-containing 16 (204341_at), score: -0.59 TRIM66tripartite motif-containing 66 (213748_at), score: -0.53 TSNtranslin (201504_s_at), score: -0.54 TSPAN13tetraspanin 13 (217979_at), score: 0.42 TSPAN32tetraspanin 32 (220558_x_at), score: 0.36 TSPYL1TSPY-like 1 (221493_at), score: 0.52 TSPYL5TSPY-like 5 (213122_at), score: 0.61 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: 0.44 TUBB2Btubulin, beta 2B (214023_x_at), score: -0.5 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.54 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.54 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: 0.48 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: -0.56 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.51 UBE2Oubiquitin-conjugating enzyme E2O (218141_at), score: 0.35 UBL3ubiquitin-like 3 (201535_at), score: 0.5 UBXN6UBX domain protein 6 (220757_s_at), score: 0.35 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: 0.57 UGCGUDP-glucose ceramide glucosyltransferase (204881_s_at), score: -0.57 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.84 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.86 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.84 UROSuroporphyrinogen III synthase (203031_s_at), score: 0.5 USP18ubiquitin specific peptidase 18 (219211_at), score: -0.59 USP25ubiquitin specific peptidase 25 (220419_s_at), score: 0.49 USP4ubiquitin specific peptidase 4 (proto-oncogene) (211800_s_at), score: 0.35 USP9Yubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) (206624_at), score: 0.36 UTP14CUTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) (203614_at), score: 0.38 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.46 VLDLRvery low density lipoprotein receptor (209822_s_at), score: 0.48 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.54 WDR48WD repeat domain 48 (222157_s_at), score: -0.59 WDR91WD repeat domain 91 (218971_s_at), score: -0.64 WNK1WNK lysine deficient protein kinase 1 (211993_at), score: -0.54 WWC3WWC family member 3 (219520_s_at), score: 0.44 WWTR1WW domain containing transcription regulator 1 (202132_at), score: 0.39 XISTX (inactive)-specific transcript (non-protein coding) (214218_s_at), score: -0.53 YIPF2Yip1 domain family, member 2 (65086_at), score: 0.38 YTHDC1YTH domain containing 1 (212455_at), score: -0.53 ZBTB43zinc finger and BTB domain containing 43 (204180_s_at), score: -0.51 ZBTB7Azinc finger and BTB domain containing 7A (219186_at), score: 0.38 ZNF131zinc finger protein 131 (214741_at), score: 0.39 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.42 ZNF224zinc finger protein 224 (216983_s_at), score: -0.63 ZNF227zinc finger protein 227 (217403_s_at), score: -0.6 ZNF230zinc finger protein 230 (205791_x_at), score: -0.52 ZNF281zinc finger protein 281 (218401_s_at), score: 0.53 ZNF331zinc finger protein 331 (219228_at), score: -0.55 ZNF415zinc finger protein 415 (205514_at), score: 0.4 ZNF562zinc finger protein 562 (219163_at), score: 0.34 ZNF654zinc finger protein 654 (219239_s_at), score: 0.43
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |