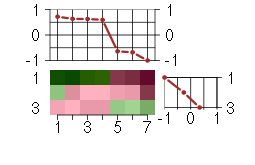

Under-expression is coded with green,
over-expression with red color.



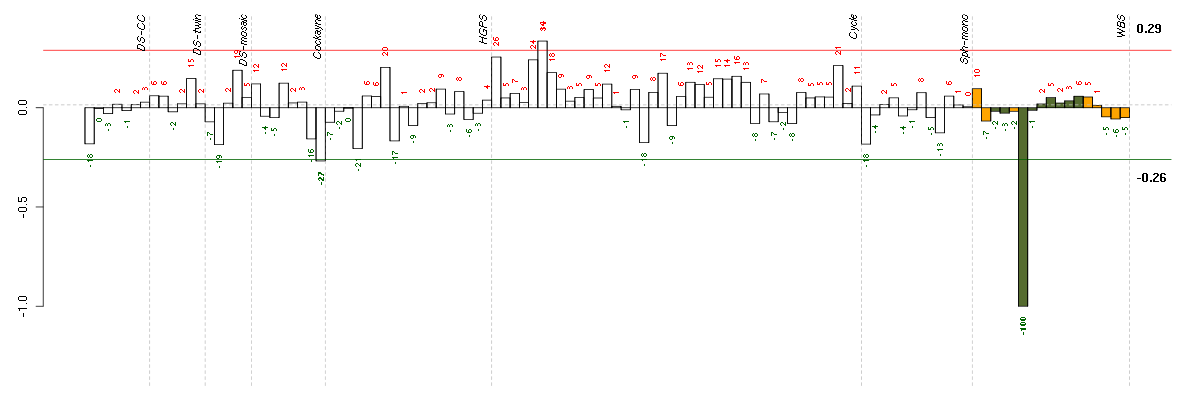
FAM115Afamily with sequence similarity 115, member A (212981_s_at), score: 0.63 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.68 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: -0.64 HIRAHIR histone cell cycle regulation defective homolog A (S. cerevisiae) (217427_s_at), score: 0.72 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: 0.63 PDIA3protein disulfide isomerase family A, member 3 (208612_at), score: 0.6 RUNDC3BRUN domain containing 3B (215321_at), score: -1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |