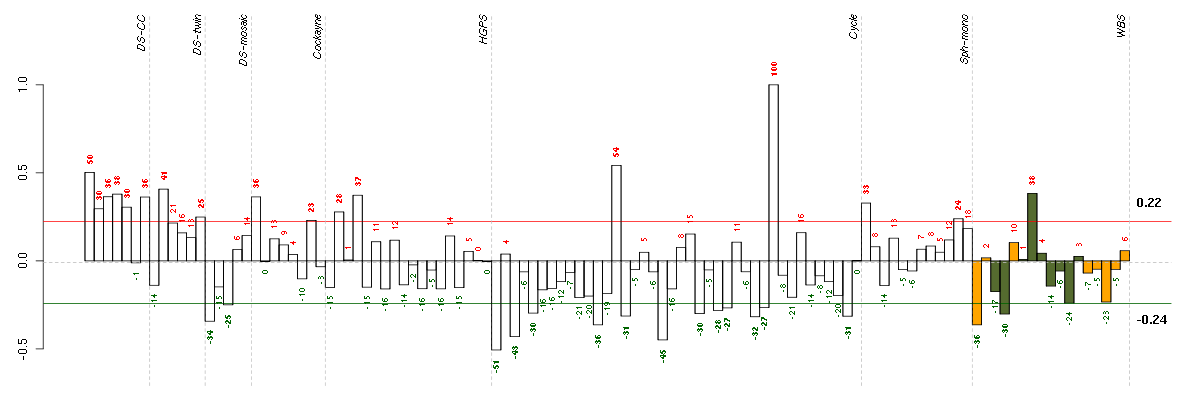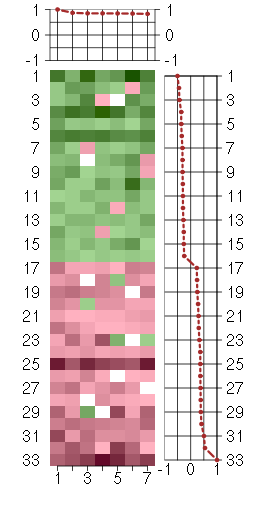

Under-expression is coded with green,
over-expression with red color.



ATRNL1attractin-like 1 (213744_at), score: 0.83 CLDN10claudin 10 (205328_at), score: 1 DSC2desmocollin 2 (204751_x_at), score: 0.84 KRT2keratin 2 (207908_at), score: 0.85 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.85 SIRPB1signal-regulatory protein beta 1 (206934_at), score: 0.85 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: 0.87
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |