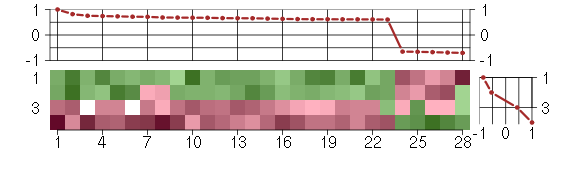

Under-expression is coded with green,
over-expression with red color.



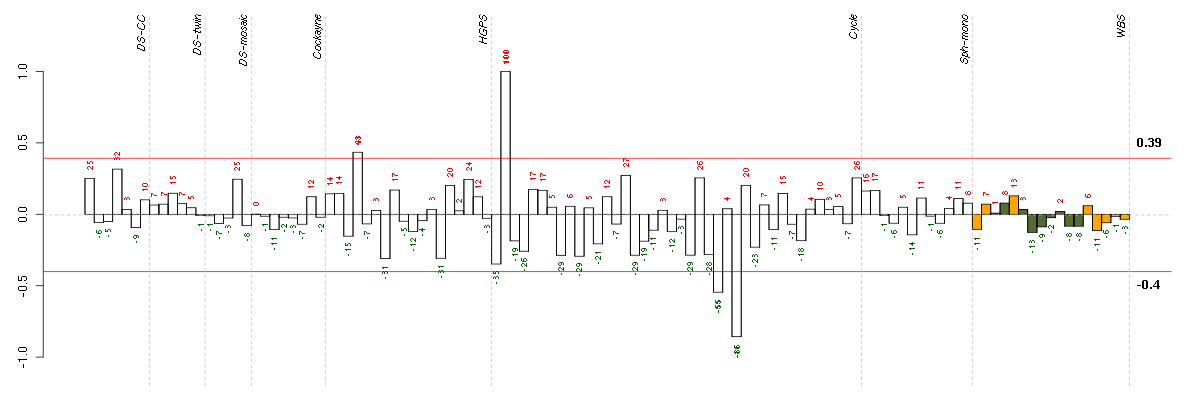
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 1 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.63 AOC2amine oxidase, copper containing 2 (retina-specific) (207064_s_at), score: 0.82 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.75 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: 0.6 FAM70Afamily with sequence similarity 70, member A (219895_at), score: -0.7 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.72 FOXO4forkhead box O4 (205451_at), score: 0.68 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: 0.74 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.71 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.62 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.69 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: 0.61 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: -0.65 MADDMAP-kinase activating death domain (38398_at), score: -0.66 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.62 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.68 NPY2Rneuropeptide Y receptor Y2 (210730_s_at), score: 0.73 POU6F1POU class 6 homeobox 1 (205878_at), score: 0.61 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.67 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.66 STXBP2syntaxin binding protein 2 (209367_at), score: 0.62 SYMPKsymplekin (202339_at), score: 0.66 SYT5synaptotagmin V (206161_s_at), score: 0.66 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.62 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.68 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.64 ZNF510zinc finger protein 510 (206053_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |