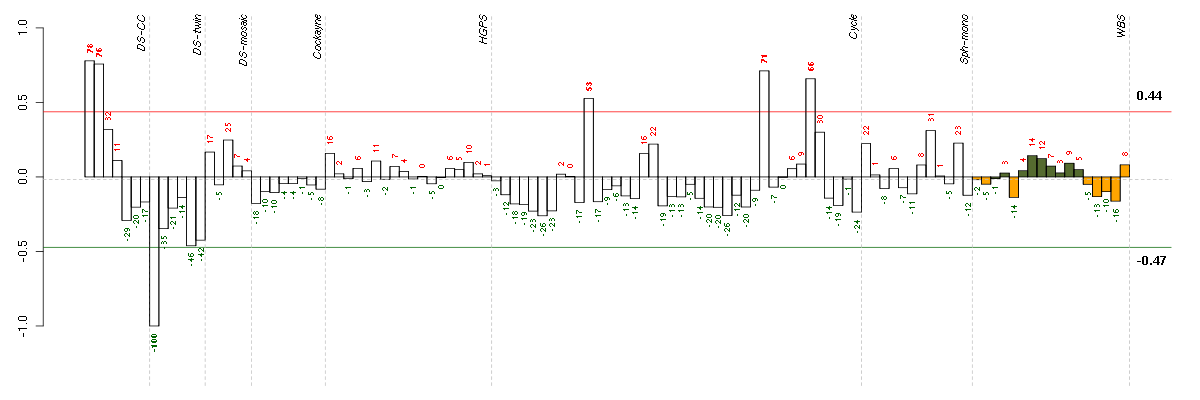

Under-expression is coded with green,
over-expression with red color.

behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 9.572e-03 | 0.5371 | 5 | 122 | Cytokine-cytokine receptor interaction |
ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.63 ANGPTL4angiopoietin-like 4 (221009_s_at), score: 0.7 BMP2bone morphogenetic protein 2 (205289_at), score: 0.58 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: 0.58 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.7 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: 0.79 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.61 EGR3early growth response 3 (206115_at), score: 0.66 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.6 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: 1 FOXO3Bforkhead box O3B pseudogene (210655_s_at), score: 0.64 GEMGTP binding protein overexpressed in skeletal muscle (204472_at), score: 0.59 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.66 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.59 INSIG1insulin induced gene 1 (201627_s_at), score: 0.71 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.7 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: 0.93 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.68 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: 0.65 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: 0.81 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: 0.76 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.79 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: 0.72 PPLperiplakin (203407_at), score: 0.58 RRADRas-related associated with diabetes (204803_s_at), score: 0.83 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: 0.59 SMOXspermine oxidase (210357_s_at), score: 0.62 THBDthrombomodulin (203887_s_at), score: 0.71 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: 0.59 VEGFAvascular endothelial growth factor A (211527_x_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |