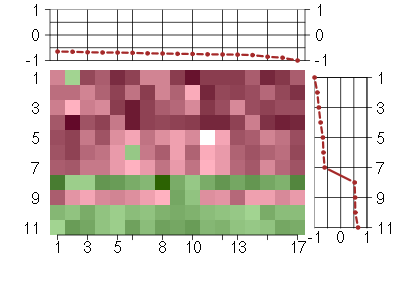

Under-expression is coded with green,
over-expression with red color.



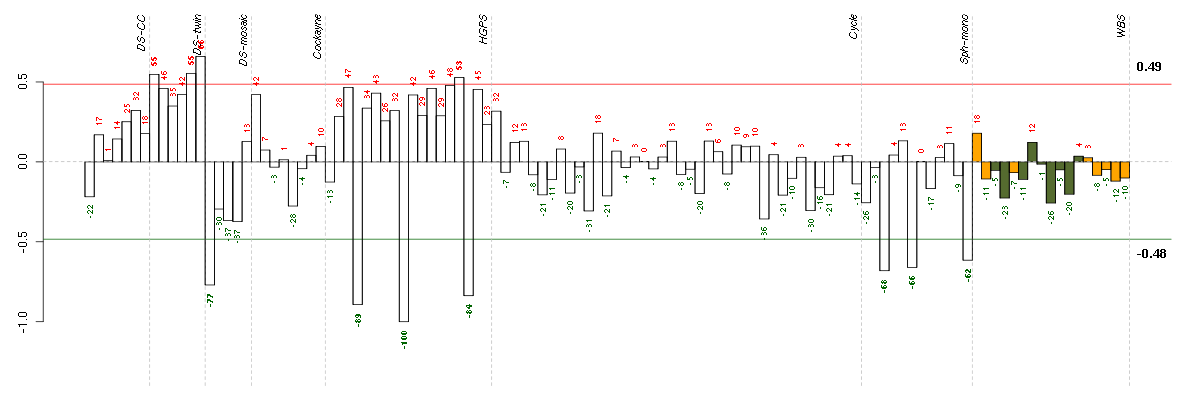
CORINcorin, serine peptidase (220356_at), score: -0.85 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: -0.72 DENND3DENN/MADD domain containing 3 (212975_at), score: -0.73 EFHD1EF-hand domain family, member D1 (209343_at), score: -0.89 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: -1 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: -0.68 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.66 LPPR4plasticity related gene 1 (213496_at), score: -0.79 MANSC1MANSC domain containing 1 (220945_x_at), score: -0.65 NRXN3neurexin 3 (205795_at), score: -0.76 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: -0.74 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: -0.7 PRSS3protease, serine, 3 (207463_x_at), score: -0.69 RTN1reticulon 1 (203485_at), score: -0.76 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.69 SYNMsynemin, intermediate filament protein (212730_at), score: -0.77 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: -0.74
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |