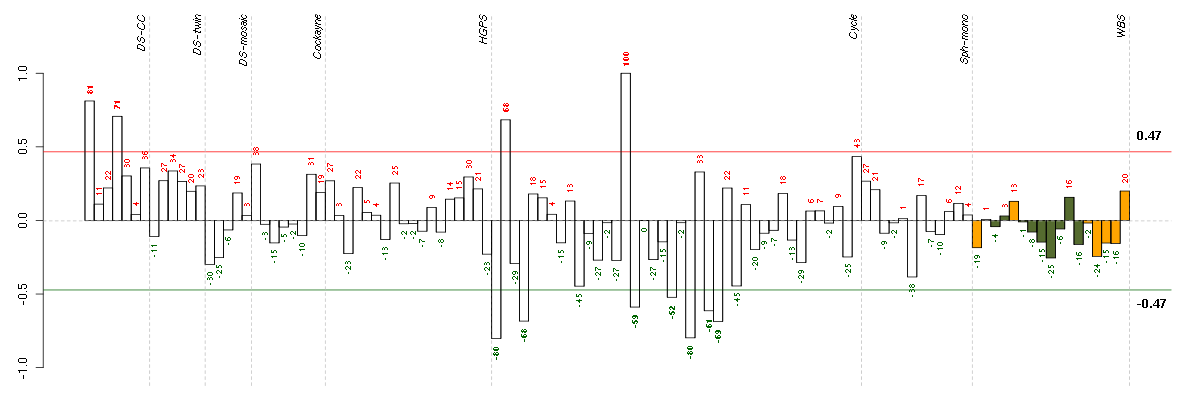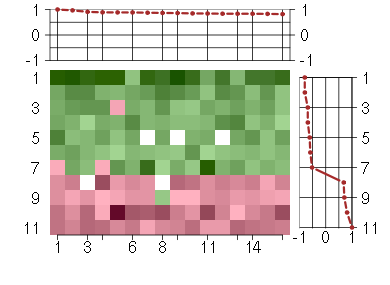

Under-expression is coded with green,
over-expression with red color.



protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
MAP-kinase scaffold activity
Functions as a physical support for the assembly of a multiprotein mitogen-activated protein kinase (MAPK) complex. MAPK scaffold proteins have binding sites for MAPK pathway kinases as well as for upstream signaling proteins.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
receptor signaling complex scaffold activity
Functions to provide a physical support for the assembly of a multiprotein receptor signaling complex.
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
all
This term is the most general term possible
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.84 EDAectodysplasin A (211130_x_at), score: 1 FBRSfibrosin (218255_s_at), score: 0.87 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.89 HAB1B1 for mucin (215778_x_at), score: 0.88 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.83 LOC80054hypothetical LOC80054 (220465_at), score: 0.86 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.84 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.83 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: 0.82 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.83 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.91 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.85 TBX21T-box 21 (220684_at), score: 0.87 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.97 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |