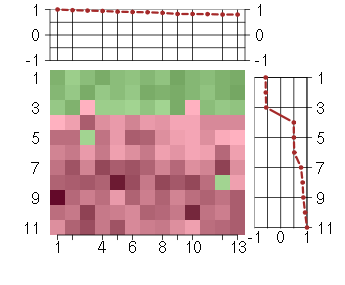

Under-expression is coded with green,
over-expression with red color.



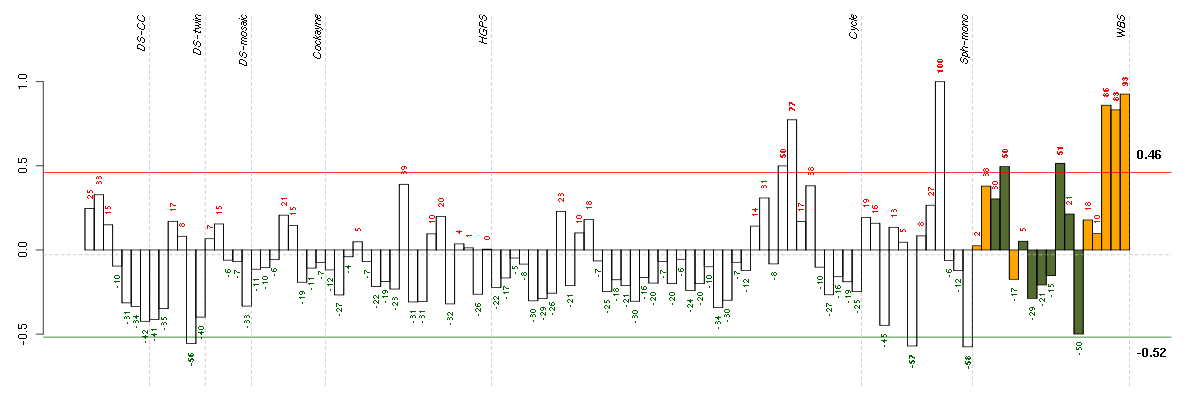
AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.8 CTSOcathepsin O (203758_at), score: 0.97 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.82 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.9 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.82 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.89 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.94 N4BP2L1NEDD4 binding protein 2-like 1 (213375_s_at), score: 0.96 SLC46A3solute carrier family 46, member 3 (214719_at), score: 1 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.87 TSC22D1TSC22 domain family, member 1 (215111_s_at), score: 0.8 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.82 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: 0.91
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |