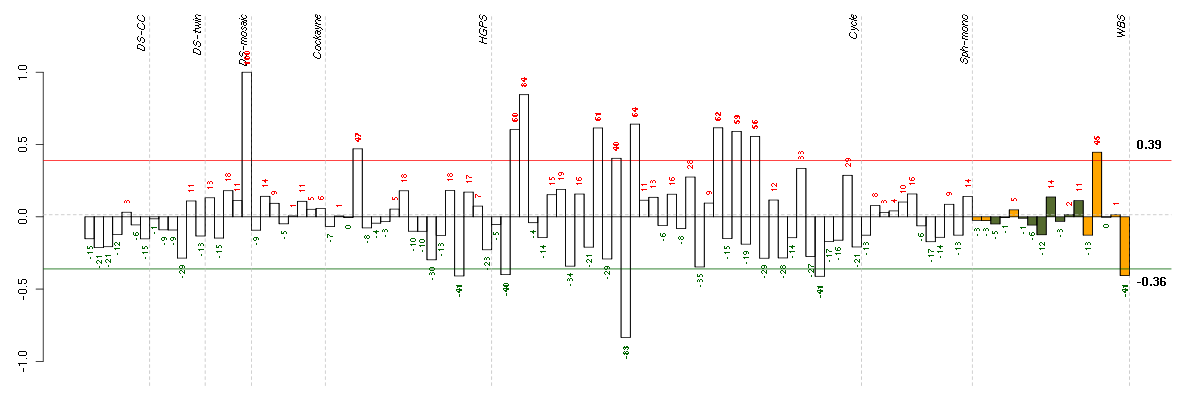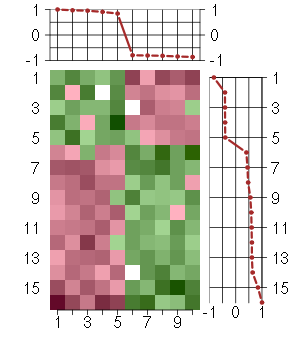

Under-expression is coded with green,
over-expression with red color.



CHMLchoroideremia-like (Rab escort protein 2) (206079_at), score: 0.84 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.82 IL1R1interleukin 1 receptor, type I (202948_at), score: -0.8 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.97 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.86 LOC391132similar to hCG2041276 (216177_at), score: -0.8 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.84 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.95 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.91 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |