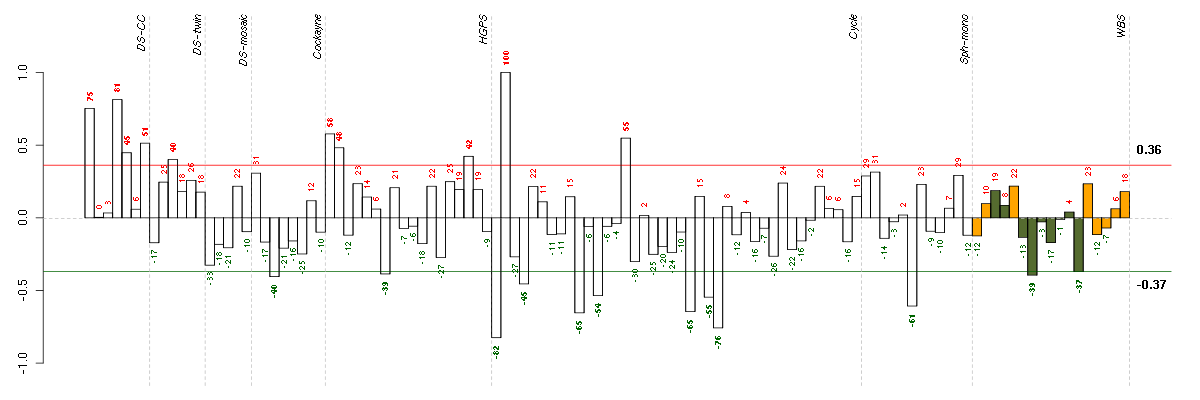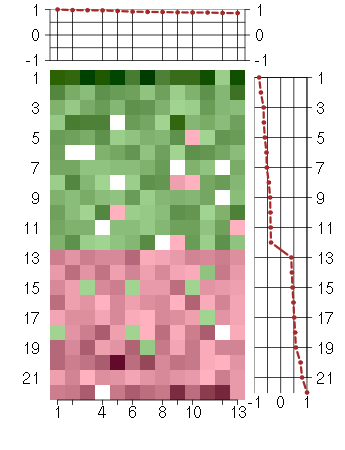

Under-expression is coded with green,
over-expression with red color.



ADORA1adenosine A1 receptor (216220_s_at), score: 0.9 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.88 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.86 AZGP1alpha-2-glycoprotein 1, zinc-binding (217014_s_at), score: 1 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.89 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.98 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.88 LOC80054hypothetical LOC80054 (220465_at), score: 0.97 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: 0.92 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.91 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.86 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.94 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.96
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| 9837_CNTL.CEL | 12 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |