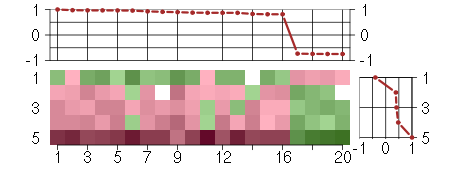

Under-expression is coded with green,
over-expression with red color.


cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cilium
A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

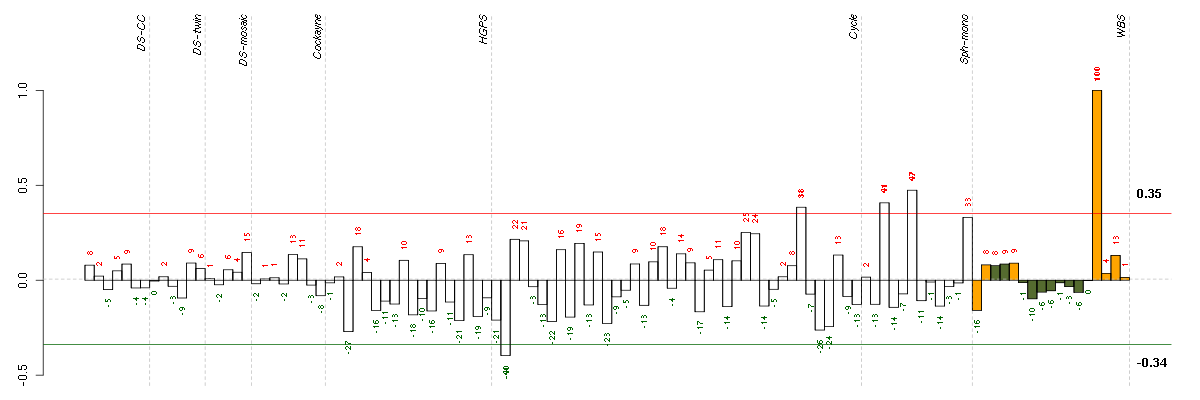
ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: -0.74 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.74 CABYRcalcium binding tyrosine-(Y)-phosphorylation regulated (219928_s_at), score: 0.97 CEP250centrosomal protein 250kDa (209495_at), score: 0.81 DKFZp547G183hypothetical LOC55525 (220572_at), score: -0.73 FAR2fatty acyl CoA reductase 2 (220615_s_at), score: 0.91 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.97 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.9 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: -0.75 KIAA1305KIAA1305 (220911_s_at), score: 0.97 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (213880_at), score: 0.87 MATN3matrilin 3 (206091_at), score: 0.87 MYRIPmyosin VIIA and Rab interacting protein (214156_at), score: 0.96 NESnestin (218678_at), score: 0.96 OLFML1olfactomedin-like 1 (217525_at), score: 0.93 SUOXsulfite oxidase (204067_at), score: 0.81 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.87 TMSB15Athymosin beta 15a (205347_s_at), score: 0.88 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.82 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |