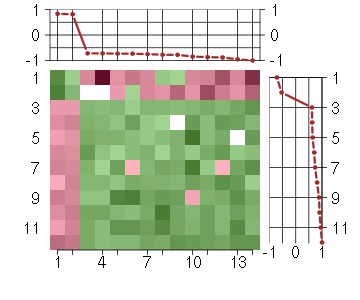

Under-expression is coded with green,
over-expression with red color.



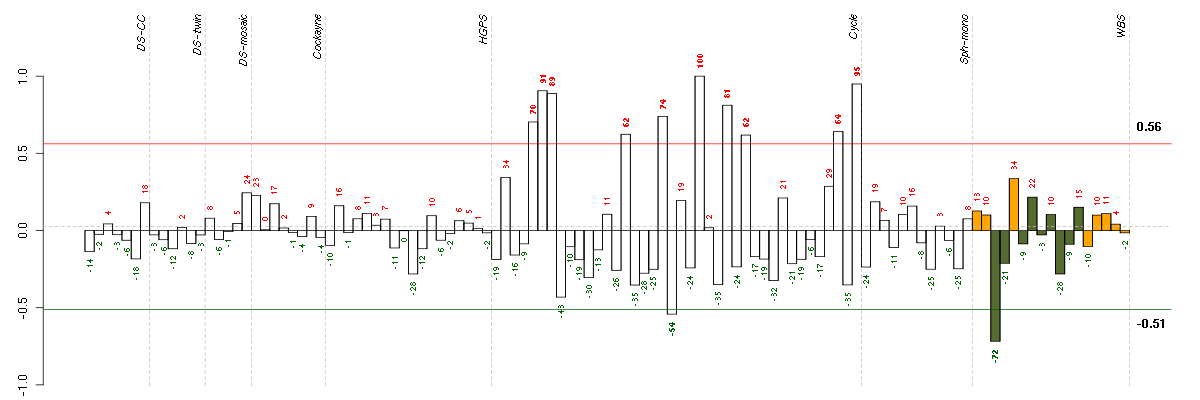
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: -0.72 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.75 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -1 F11coagulation factor XI (206610_s_at), score: 0.83 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.73 FOXK2forkhead box K2 (203064_s_at), score: -0.95 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.77 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.81 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.87 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: -0.73 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.85 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.78 SETBP1SET binding protein 1 (205933_at), score: -0.72 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: -0.88
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |