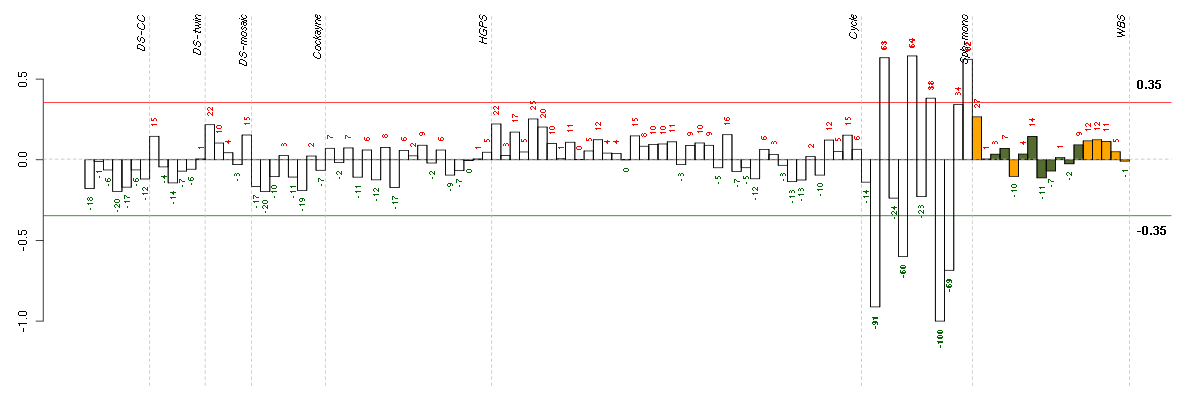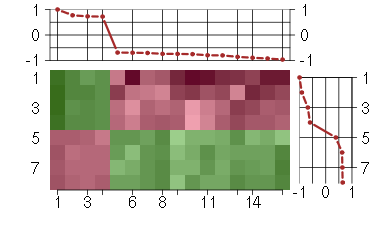

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00120 | 2.491e-02 | 0.03118 | 2 | 17 | Bile acid biosynthesis |
| 00650 | 4.768e-02 | 0.04769 | 2 | 26 | Butanoate metabolism |
AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.92 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.7 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.72 C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.74 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.8 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.74 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.75 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.86 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.96 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.79 LUMlumican (201744_s_at), score: 0.73 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.89 MYO1Bmyosin IB (212364_at), score: 0.77 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.7 SPANXCSPANX family, member C (220217_x_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |