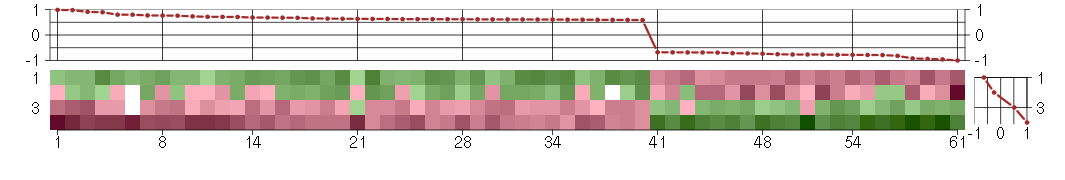

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

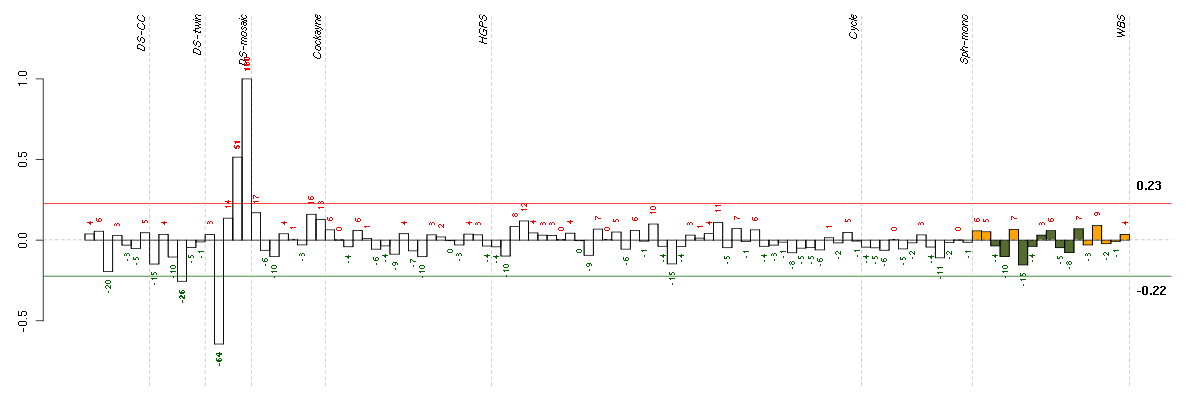
ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.76 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.63 B9D1B9 protein domain 1 (210534_s_at), score: -0.68 BAT2D1BAT2 domain containing 1 (211947_s_at), score: 0.61 BCHEbutyrylcholinesterase (205433_at), score: 0.91 C10orf10chromosome 10 open reading frame 10 (209182_s_at), score: 0.61 C1orf38chromosome 1 open reading frame 38 (207571_x_at), score: -0.69 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.68 CFBcomplement factor B (202357_s_at), score: -0.77 CFIcomplement factor I (203854_at), score: -0.94 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.59 CYTH3cytohesin 3 (206523_at), score: 0.62 CYTIPcytohesin 1 interacting protein (209606_at), score: 0.97 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.62 DSC2desmocollin 2 (204751_x_at), score: 0.69 ECHDC2enoyl Coenzyme A hydratase domain containing 2 (218552_at), score: -0.71 EDN1endothelin 1 (218995_s_at), score: 0.64 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.78 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: -0.82 FN1fibronectin 1 (214701_s_at), score: 0.62 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: -0.77 GCH1GTP cyclohydrolase 1 (204224_s_at), score: -0.78 GPC4glypican 4 (204983_s_at), score: -0.79 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.61 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: 0.62 HOXB9homeobox B9 (216417_x_at), score: 0.98 INHBEinhibin, beta E (210587_at), score: -1 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.73 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: 0.63 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.6 MACROD1MACRO domain containing 1 (219188_s_at), score: -0.75 MAXMYC associated factor X (210734_x_at), score: 0.63 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.61 MFN2mitofusin 2 (216205_s_at), score: 0.67 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.76 MTMR1myotubularin related protein 1 (214975_s_at), score: 0.64 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.71 PAPPA2pappalysin 2 (213332_at), score: -0.77 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: 0.71 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.61 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.6 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.59 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: -0.68 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.71 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.73 PTRFpolymerase I and transcript release factor (208790_s_at), score: 0.61 PXNpaxillin (211823_s_at), score: 0.6 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.96 SEC14L1SEC14-like 1 (S. cerevisiae) (202082_s_at), score: 0.65 SLC26A6solute carrier family 26, member 6 (221572_s_at), score: -0.92 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.77 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.68 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.59 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.89 SULT1A3sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (210580_x_at), score: -0.68 TANC2tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 (208425_s_at), score: 0.8 TAPBPLTAP binding protein-like (218746_at), score: -0.78 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: 0.62 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.8 ZNF467zinc finger protein 467 (214746_s_at), score: -0.72 ZNF688zinc finger protein 688 (213527_s_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |