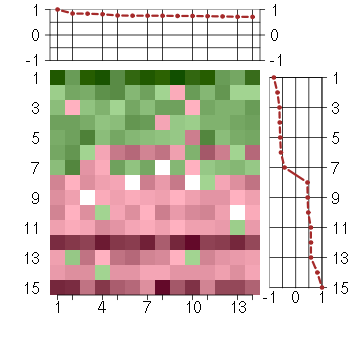

Under-expression is coded with green,
over-expression with red color.


cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

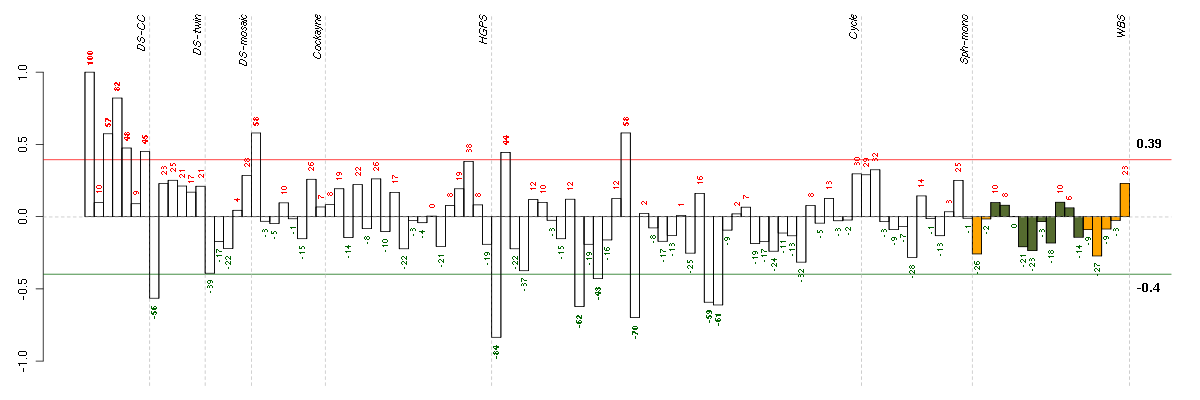
C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.73 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.71 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.74 EDAectodysplasin A (211130_x_at), score: 0.84 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.71 MMRN2multimerin 2 (219091_s_at), score: 0.75 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.75 MYO1Amyosin IA (211916_s_at), score: 0.81 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: 0.84 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.77 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 1 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.76 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.76 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |