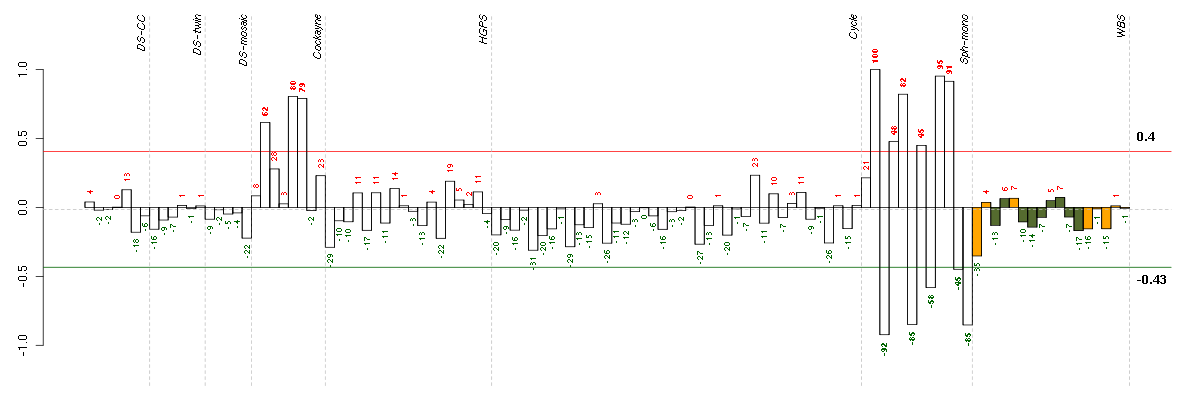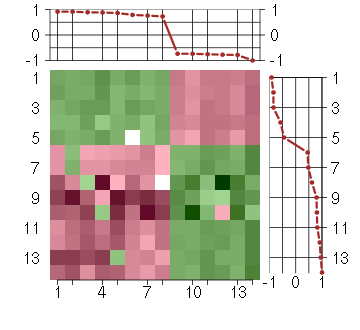

Under-expression is coded with green,
over-expression with red color.



AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.91 CD24CD24 molecule (209771_x_at), score: 0.73 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.79 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.89 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.88 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.92 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.74 LUMlumican (201744_s_at), score: -0.77 MYO1Bmyosin IB (212364_at), score: -0.75 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.86 TNS1tensin 1 (221748_s_at), score: -0.78 TSPYL5TSPY-like 5 (213122_at), score: -0.73 WT1Wilms tumor 1 (206067_s_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |