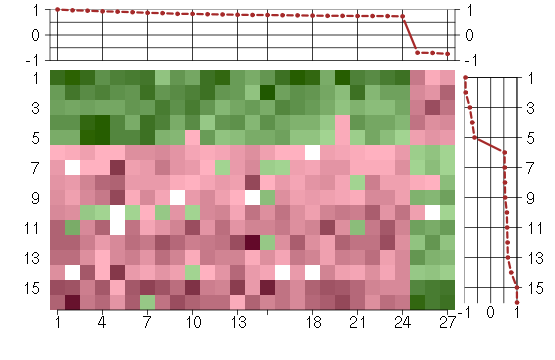

Under-expression is coded with green,
over-expression with red color.



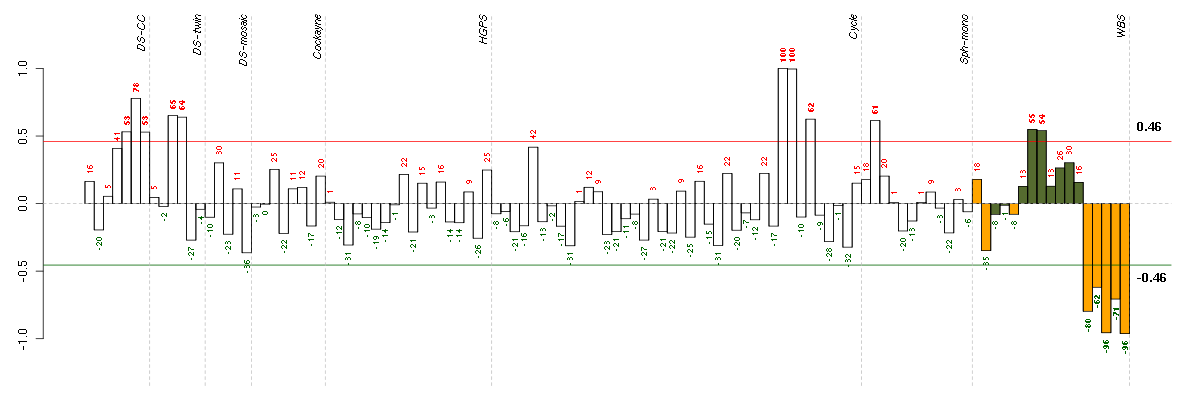
ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.75 ARNTL2aryl hydrocarbon receptor nuclear translocator-like 2 (220658_s_at), score: -0.74 ASNSasparagine synthetase (205047_s_at), score: 0.73 ATF4activating transcription factor 4 (tax-responsive enhancer element B67) (200779_at), score: 0.83 C9orf91chromosome 9 open reading frame 91 (221865_at), score: 0.74 CEBPGCCAAT/enhancer binding protein (C/EBP), gamma (204203_at), score: 0.81 DAPK1death-associated protein kinase 1 (203139_at), score: 0.83 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 0.97 DDIT4DNA-damage-inducible transcript 4 (202887_s_at), score: 0.74 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.7 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.87 FAM129Afamily with sequence similarity 129, member A (217966_s_at), score: 0.78 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.93 FLGfilaggrin (215704_at), score: 0.91 FMODfibromodulin (202709_at), score: 0.86 GDF15growth differentiation factor 15 (221577_x_at), score: 0.89 GOLSYNGolgi-localized protein (218692_at), score: 0.95 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (217168_s_at), score: 0.74 INHBEinhibin, beta E (210587_at), score: 0.79 PALMparalemmin (203859_s_at), score: 0.79 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (202847_at), score: 0.76 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.81 SLC3A2solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 (200924_s_at), score: 0.77 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.78 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.69 TRIB3tribbles homolog 3 (Drosophila) (218145_at), score: 1 TSC22D3TSC22 domain family, member 3 (208763_s_at), score: 0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 5850_CNTL.CEL | 8 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |