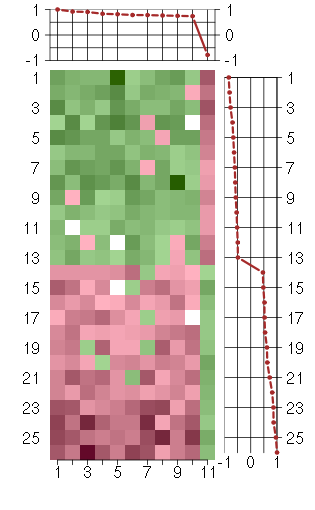

Under-expression is coded with green,
over-expression with red color.



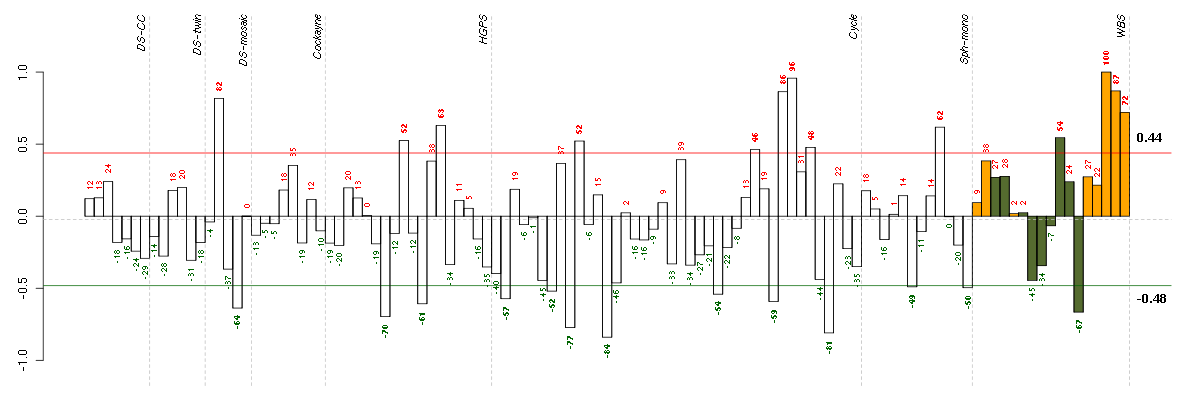
ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.78 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.83 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.78 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.92 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.74 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.76 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.78 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.75 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.9 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| 9837_CNTL.CEL | 12 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |