
Under-expression is coded with green,
over-expression with red color.

response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

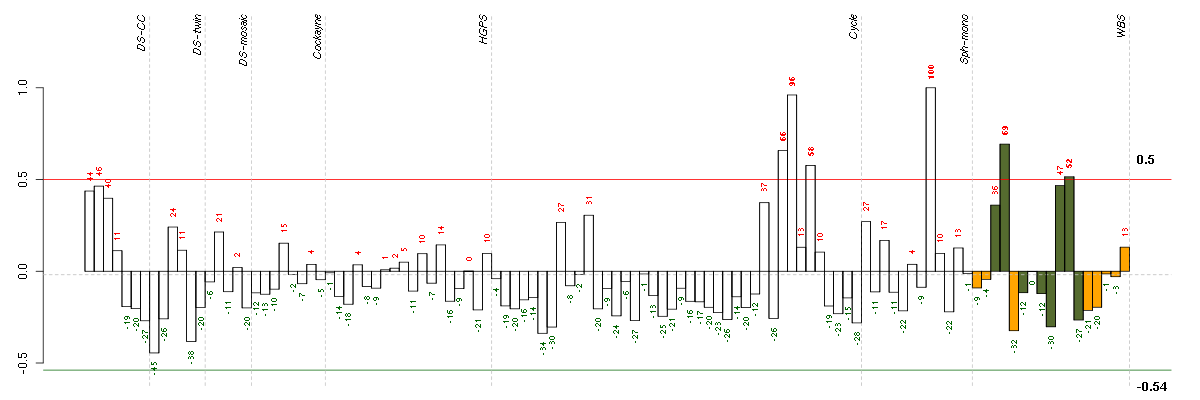
ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.73 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.74 ANK2ankyrin 2, neuronal (202920_at), score: 0.67 BDKRB2bradykinin receptor B2 (205870_at), score: 0.76 BTG2BTG family, member 2 (201236_s_at), score: 0.65 C3complement component 3 (217767_at), score: 0.73 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: 0.66 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 1 CCL8chemokine (C-C motif) ligand 8 (214038_at), score: 0.82 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.7 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.7 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (221892_at), score: 0.66 IRAK3interleukin-1 receptor-associated kinase 3 (213817_at), score: 0.84 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.78 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: 0.65 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.72 PDPNpodoplanin (221898_at), score: 0.82 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.65 PPLperiplakin (203407_at), score: 0.82 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: 0.65 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.7 SESN1sestrin 1 (218346_s_at), score: 0.76 SLC1A3solute carrier family 1 (glial high affinity glutamate transporter), member 3 (202800_at), score: 0.94 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.69 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.73
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |