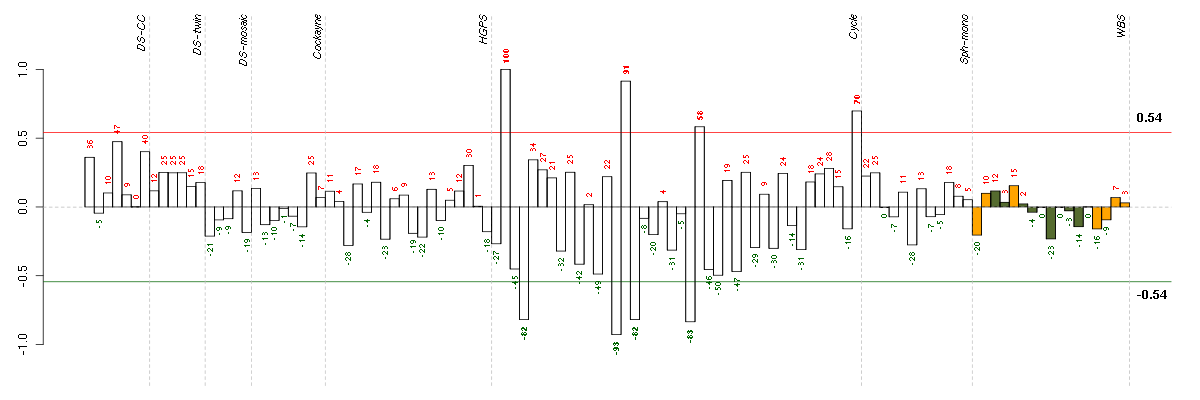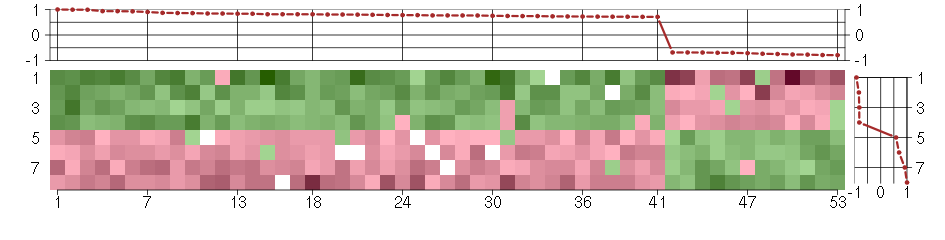

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.81 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.68 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.78 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.75 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.73 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.75 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.84 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.72 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.71 DSPPdentin sialophosphoprotein (221681_s_at), score: 1 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.72 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.68 FBRSfibrosin (218255_s_at), score: 0.81 FBXO2F-box protein 2 (219305_x_at), score: 0.86 FGL1fibrinogen-like 1 (205305_at), score: 0.77 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.78 FKRPfukutin related protein (219853_at), score: -0.77 FOXK2forkhead box K2 (203064_s_at), score: -0.79 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: 0.71 GPR144G protein-coupled receptor 144 (216289_at), score: 0.99 HAB1B1 for mucin (215778_x_at), score: 0.93 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.77 HCG8HLA complex group 8 (215985_at), score: -0.76 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.8 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: 0.75 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.9 KIF26Bkinesin family member 26B (220002_at), score: 0.72 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.87 LOC149501similar to keratin 8 (216821_at), score: 0.93 LOC80054hypothetical LOC80054 (220465_at), score: 0.81 MATN3matrilin 3 (206091_at), score: -0.74 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.74 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.8 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.7 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.72 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.94 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.77 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.86 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: 0.81 RBM12BRNA binding motif protein 12B (51228_at), score: -0.69 RBM38RNA binding motif protein 38 (212430_at), score: 0.84 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.74 SAA4serum amyloid A4, constitutive (207096_at), score: 0.79 SEPT5septin 5 (209767_s_at), score: 0.78 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.99 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.83 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.7 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.8 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.83 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.73 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.76 TWISTNBTWIST neighbor (214729_at), score: -0.79 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |