

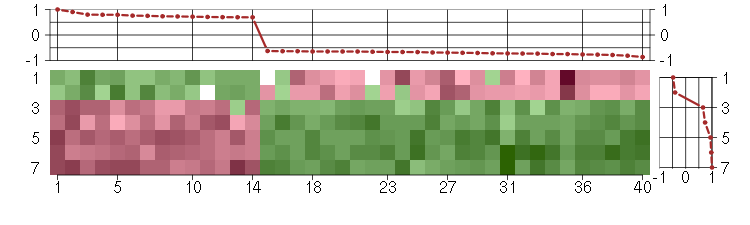

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ANXA10annexin A10 (210143_at), score: -0.7 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.7 CD320CD320 molecule (218529_at), score: 0.73 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.67 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.75 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.77 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.81 CST6cystatin E/M (206595_at), score: -0.64 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.76 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.64 DUSP6dual specificity phosphatase 6 (208892_s_at), score: -0.69 EMCNendomucin (219436_s_at), score: 0.8 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.73 GAAglucosidase, alpha; acid (202812_at), score: -0.65 GPM6Bglycoprotein M6B (209170_s_at), score: -0.73 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.79 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.67 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.65 IL33interleukin 33 (209821_at), score: -0.86 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.69 KRT33Akeratin 33A (208483_x_at), score: -0.65 MLPHmelanophilin (218211_s_at), score: 0.72 NEFLneurofilament, light polypeptide (221805_at), score: -0.75 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.7 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.63 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204286_s_at), score: -0.66 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.72 PRSS3protease, serine, 3 (207463_x_at), score: -0.78 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: -0.72 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.7 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.65 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203788_s_at), score: -0.69 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.79 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.71 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.9 SYT11synaptotagmin XI (209198_s_at), score: 0.75 TMSB15Athymosin beta 15a (205347_s_at), score: 0.76 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.73 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 ZNF219zinc finger protein 219 (219314_s_at), score: -0.67
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
