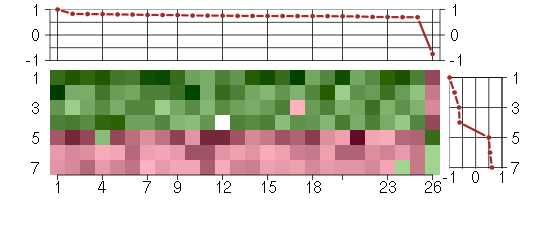

Under-expression is coded with green,
over-expression with red color.



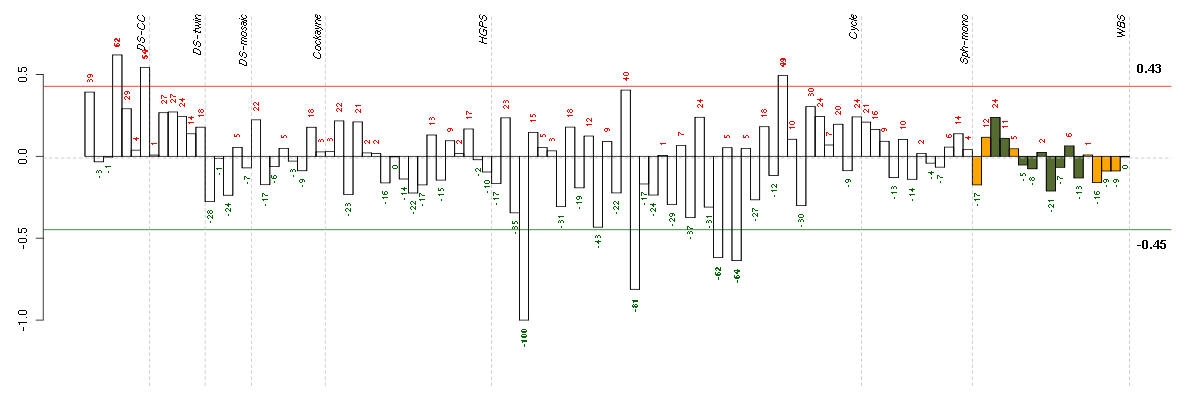
ADORA1adenosine A1 receptor (216220_s_at), score: 0.7 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.74 ATP6V1G2ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 (214762_at), score: 0.76 CPA4carboxypeptidase A4 (205832_at), score: 0.74 DOK4docking protein 4 (209691_s_at), score: -0.74 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.7 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.75 F11coagulation factor XI (206610_s_at), score: 0.76 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.7 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.74 GPR144G protein-coupled receptor 144 (216289_at), score: 1 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: 0.78 HAB1B1 for mucin (215778_x_at), score: 0.75 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.79 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.74 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.83 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.81 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: 0.71 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.73 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.74 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.8 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.82 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.78 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.73 TYMPthymidine phosphorylase (217497_at), score: 0.82 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |