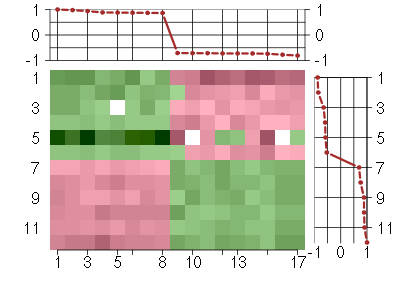

Under-expression is coded with green,
over-expression with red color.



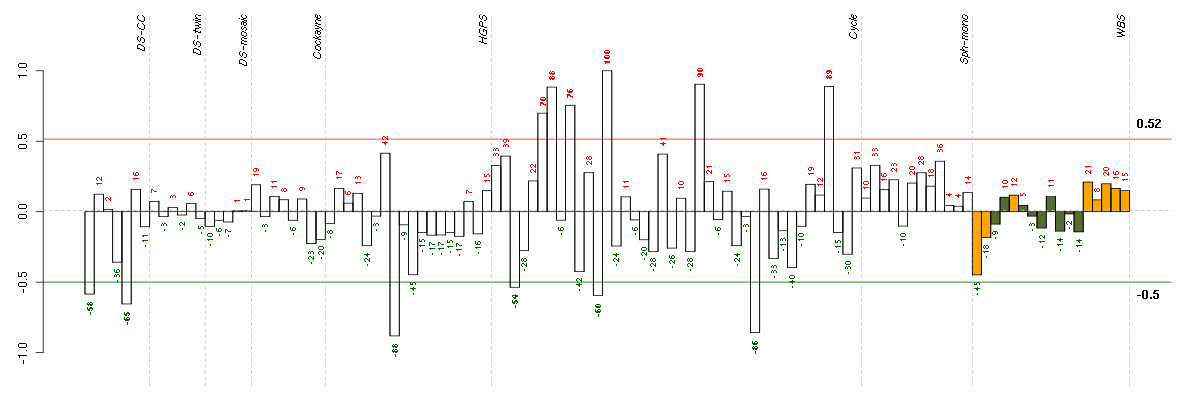
ATN1atrophin 1 (40489_at), score: -0.74 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.88 CHMP5chromatin modifying protein 5 (218085_at), score: 0.94 CICcapicua homolog (Drosophila) (212784_at), score: -0.71 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 1 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: 0.86 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.77 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.72 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.72 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.81 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.87 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.71 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.71 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.88 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.72 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.98 YTHDC2YTH domain containing 2 (213077_at), score: 0.87
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |