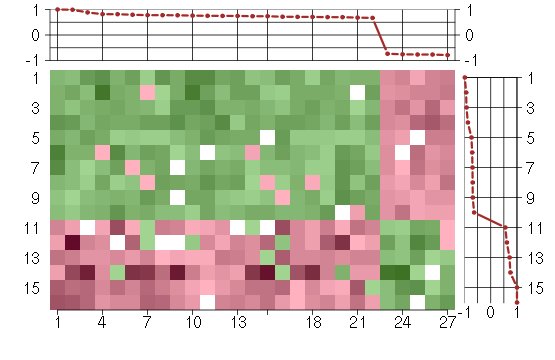

Under-expression is coded with green,
over-expression with red color.



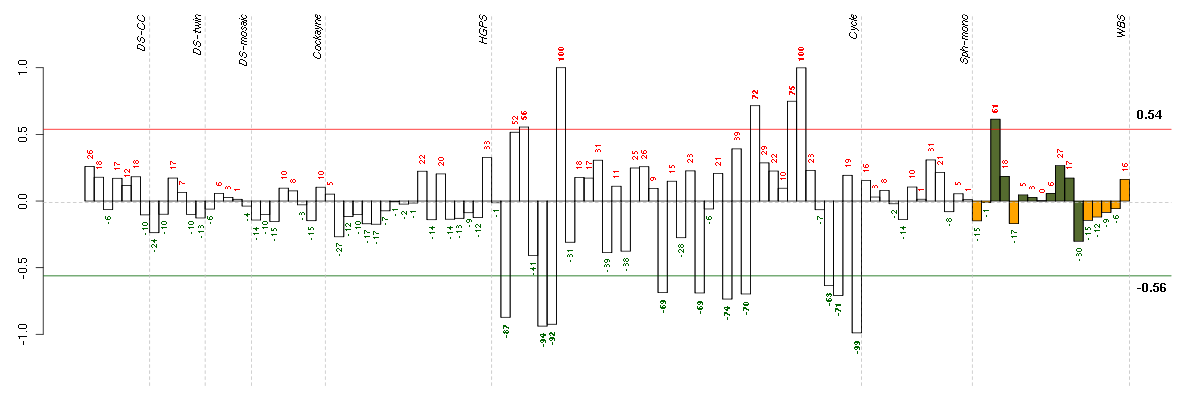
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.99 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.75 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.74 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.71 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.67 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.7 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.74 FOXK2forkhead box K2 (203064_s_at), score: 0.75 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.79 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.76 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 1 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.71 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.77 KIAA1305KIAA1305 (220911_s_at), score: 0.77 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.82 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.74 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.75 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: -0.77 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.72 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.89 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.7 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: 0.78 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.78 SYT1synaptotagmin I (203999_at), score: 0.68 TSKUtsukushin (218245_at), score: 0.82 ULBP2UL16 binding protein 2 (221291_at), score: -0.76 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |