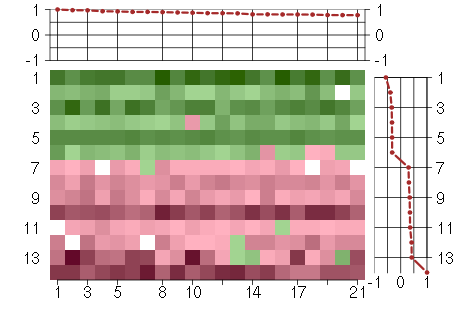

Under-expression is coded with green,
over-expression with red color.

transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
neurotransmitter transport
The directed movement of a neurotransmitter into, out of, within or between cells. Neurotransmitters are any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.


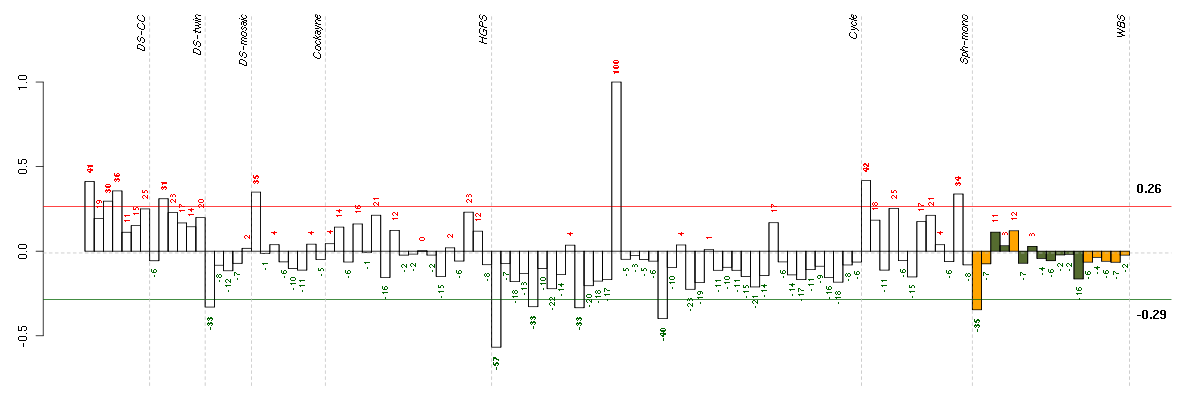
ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.92 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.78 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.93 DTNBdystrobrevin, beta (215295_at), score: 0.88 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.91 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.81 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.97 LHX3LIM homeobox 3 (221670_s_at), score: 0.8 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.91 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.81 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.97 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.78 MYO5Cmyosin VC (218966_at), score: 0.85 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.81 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: 0.85 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.9 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.87 SFTPBsurfactant protein B (214354_x_at), score: 0.8 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.86 TAAR2trace amine associated receptor 2 (221394_at), score: 1 ZNF287zinc finger protein 287 (216710_x_at), score: 0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |