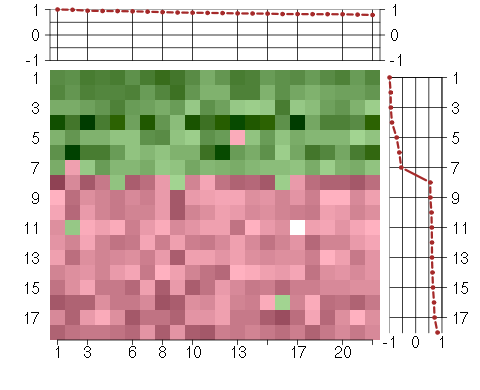

Under-expression is coded with green,
over-expression with red color.



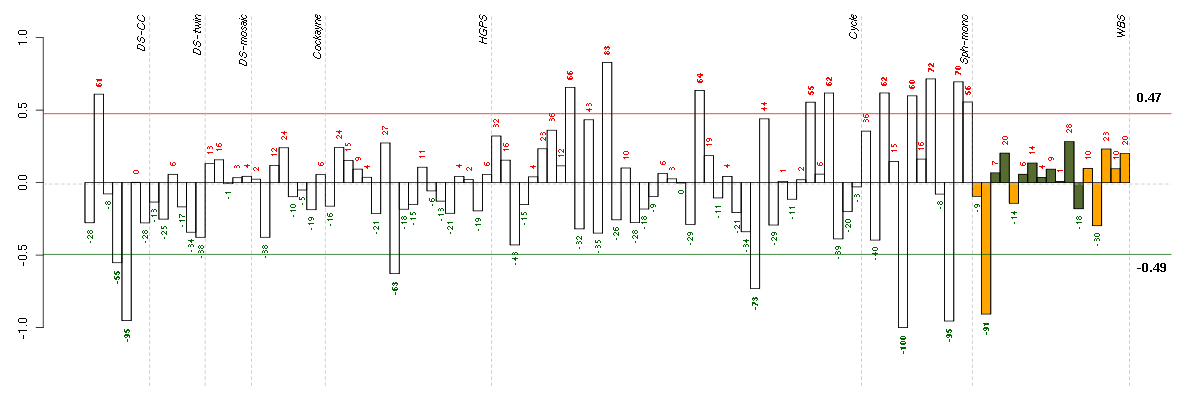
ABI1abl-interactor 1 (209028_s_at), score: 0.81 ATF1activating transcription factor 1 (222103_at), score: 0.91 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.9 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.94 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.82 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.85 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.88 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.86 JMJD1Cjumonji domain containing 1C (221763_at), score: 1 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (218311_at), score: 0.78 MYO1Bmyosin IB (212364_at), score: 0.99 MYO6myosin VI (203216_s_at), score: 0.8 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.93 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.95 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.82 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.82 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.87 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.94 TMF1TATA element modulatory factor 1 (213024_at), score: 0.87 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.84 YTHDC2YTH domain containing 2 (213077_at), score: 0.82 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |