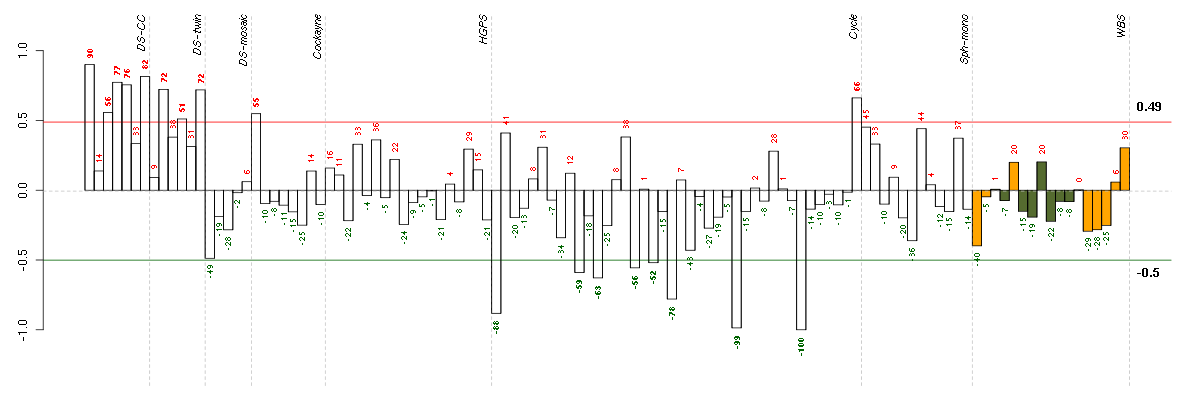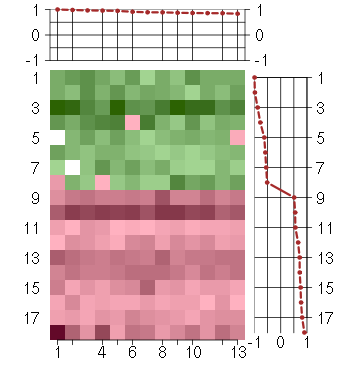

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00472 | 4.274e-02 | 0.0007337 | 1 | 1 | D-Arginine and D-ornithine metabolism |
C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.96 DAOD-amino-acid oxidase (206878_at), score: 0.95 EDARectodysplasin A receptor (220048_at), score: 0.89 GPATCH4G patch domain containing 4 (220596_at), score: 0.86 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.85 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: 0.96 PEG3paternally expressed 3 (209242_at), score: 0.89 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.91 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.98 SERPINB3serpin peptidase inhibitor, clade B (ovalbumin), member 3 (209719_x_at), score: 0.83 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.87 TRIM45tripartite motif-containing 45 (219923_at), score: 0.86 VGFVGF nerve growth factor inducible (205586_x_at), score: 1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |