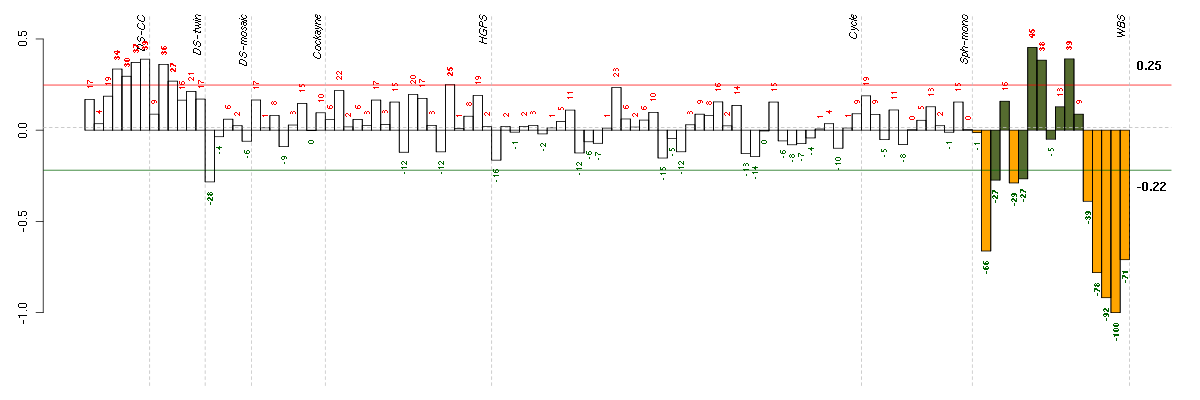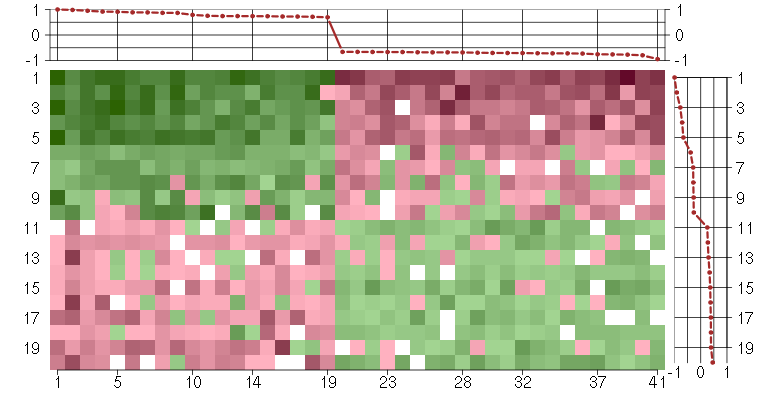

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ABHD14Aabhydrolase domain containing 14A (210006_at), score: -0.66 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.69 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: -0.67 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: -0.69 BTDbiotinidase (204167_at), score: -0.72 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: -0.76 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: 0.79 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.74 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: -0.95 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: -0.68 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.72 DBC1deleted in bladder cancer 1 (205818_at), score: 0.89 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: -0.68 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.95 ENDOD1endonuclease domain containing 1 (212573_at), score: -0.71 F10coagulation factor X (205620_at), score: -0.77 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 1 FLGfilaggrin (215704_at), score: 0.98 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.74 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: -0.67 GOLSYNGolgi-localized protein (218692_at), score: 0.92 HOXA7homeobox A7 (206848_at), score: 0.74 IGF1insulin-like growth factor 1 (somatomedin C) (209541_at), score: 0.89 ITGB8integrin, beta 8 (211488_s_at), score: 0.73 ITIH5inter-alpha (globulin) inhibitor H5 (219064_at), score: 0.74 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: -0.7 LEPREL2leprecan-like 2 (204854_at), score: -0.66 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: -0.72 LPCAT4lysophosphatidylcholine acyltransferase 4 (40472_at), score: -0.72 NME3non-metastatic cells 3, protein expressed in (204862_s_at), score: -0.8 NRCAMneuronal cell adhesion molecule (204105_s_at), score: 0.87 OCEL1occludin/ELL domain containing 1 (205441_at), score: -0.68 PAPPA2pappalysin 2 (213332_at), score: 0.71 PLGLB2plasminogen-like B2 (205871_at), score: 0.75 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.68 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.71 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.87 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.91 TAP1transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (202307_s_at), score: -0.67 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: -0.73 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 5850_CNTL.CEL | 8 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |