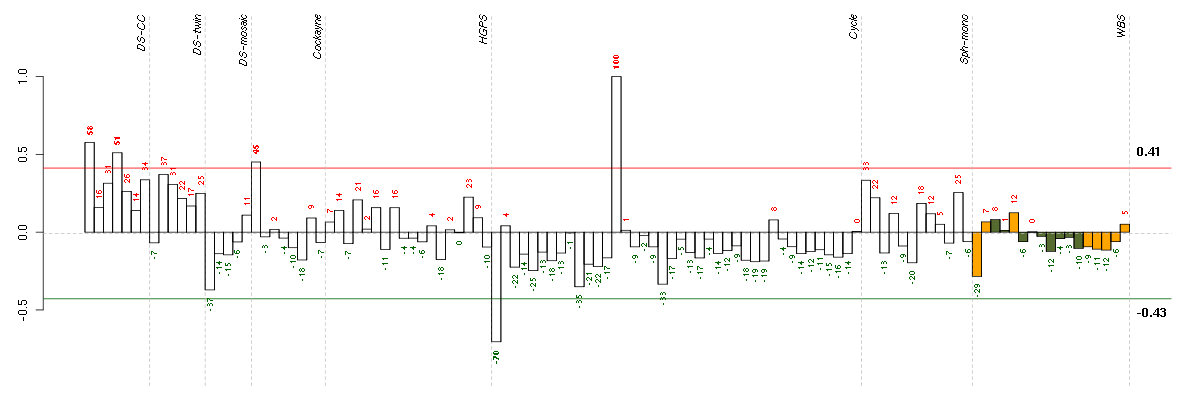

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.68 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: 0.91 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.87 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.87 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: 0.68 BEANbrain expressed, associated with Nedd4 (214068_at), score: 0.68 BMP10bone morphogenetic protein 10 (208292_at), score: 0.69 BRS3bombesin-like receptor 3 (207369_at), score: 0.77 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.77 CD28CD28 molecule (206545_at), score: 0.72 CDH19cadherin 19, type 2 (206898_at), score: 1 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: 0.8 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.86 CLDN10claudin 10 (205328_at), score: 0.76 CTSScathepsin S (202901_x_at), score: 0.67 CYCSP52cytochrome c, somatic pseudogene 52 (217206_at), score: 0.77 DHRS9dehydrogenase/reductase (SDR family) member 9 (219799_s_at), score: 0.85 DSC2desmocollin 2 (204751_x_at), score: 0.7 DTNBdystrobrevin, beta (215295_at), score: 0.88 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.73 GLRA1glycine receptor, alpha 1 (207972_at), score: 0.74 GPR12G protein-coupled receptor 12 (214558_at), score: 0.74 GPR63G protein-coupled receptor 63 (220993_s_at), score: 0.74 HCG8HLA complex group 8 (215985_at), score: 0.69 HOXB6homeobox B6 (205366_s_at), score: 0.67 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.84 KCNIP1Kv channel interacting protein 1 (221307_at), score: 0.76 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.82 KRT2keratin 2 (207908_at), score: 0.71 LHX3LIM homeobox 3 (221670_s_at), score: 0.83 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.85 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.88 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.76 MBmyoglobin (204179_at), score: 0.72 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.69 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: 0.67 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.71 MMRN2multimerin 2 (219091_s_at), score: 0.68 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.76 MYO1Amyosin IA (211916_s_at), score: 0.99 MYO5Cmyosin VC (218966_at), score: 0.87 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.82 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.7 NRN1neuritin 1 (218625_at), score: 0.7 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.81 PCDH11Yprotocadherin 11 Y-linked (211227_s_at), score: 0.82 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: 0.91 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.69 PEG3paternally expressed 3 (209242_at), score: 0.7 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.71 PRLprolactin (205445_at), score: 0.84 PROL1proline rich, lacrimal 1 (208004_at), score: 0.73 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.84 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.73 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.72 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.93 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.75 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: 0.7 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (206830_at), score: 0.75 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.78 TAAR2trace amine associated receptor 2 (221394_at), score: 0.93 TAS2R9taste receptor, type 2, member 9 (221461_at), score: 0.71 TLR1toll-like receptor 1 (210176_at), score: 0.88 TSPY1testis specific protein, Y-linked 1 (216374_at), score: 0.79 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.84 ZNF287zinc finger protein 287 (216710_x_at), score: 0.7
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |