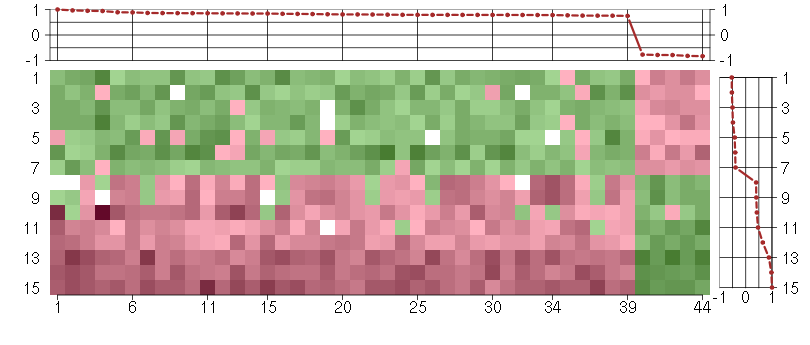

Under-expression is coded with green,
over-expression with red color.



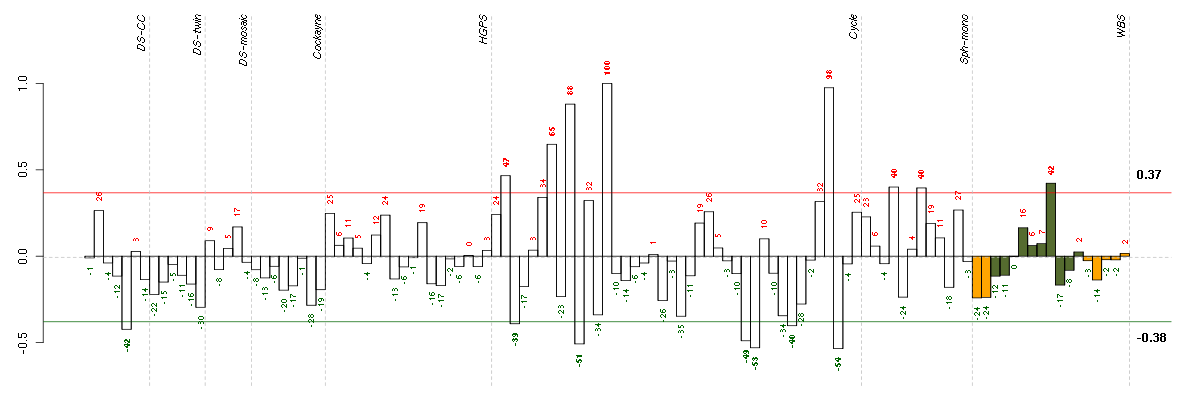
ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.76 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.85 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.81 ADORA2Badenosine A2b receptor (205891_at), score: -0.79 AHI1Abelson helper integration site 1 (221569_at), score: 0.78 AIM1absent in melanoma 1 (212543_at), score: -0.77 ANXA10annexin A10 (210143_at), score: 0.86 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.76 CASC3cancer susceptibility candidate 3 (207842_s_at), score: -0.78 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.85 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: 0.79 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.75 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.85 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.95 DUSP4dual specificity phosphatase 4 (204014_at), score: 0.82 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.77 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: 0.8 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.83 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.81 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.79 GKglycerol kinase (207387_s_at), score: 0.79 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.87 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.79 IL33interleukin 33 (209821_at), score: 0.94 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.96 KIAA0562KIAA0562 (204075_s_at), score: 0.79 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.89 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.81 NEFMneurofilament, medium polypeptide (205113_at), score: 1 NPTX1neuronal pentraxin I (204684_at), score: 0.79 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.76 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.89 PCDH9protocadherin 9 (219737_s_at), score: 0.84 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: 0.85 RNF11ring finger protein 11 (208924_at), score: 0.84 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.8 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.83 SYNJ1synaptojanin 1 (212990_at), score: 0.79 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.78 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.86 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.78 TP53tumor protein p53 (201746_at), score: -0.83 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: 0.79 WDR6WD repeat domain 6 (217734_s_at), score: -0.82
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 6089_CNTL.CEL | 9 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |