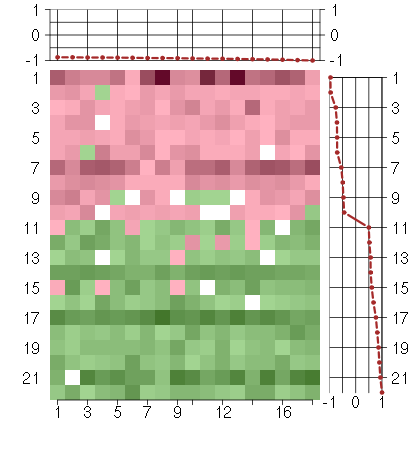

Under-expression is coded with green,
over-expression with red color.


cell fraction
A generic term for parts of cells prepared by disruptive biochemical techniques.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane fraction
That fraction of cells, prepared by disruptive biochemical methods, that includes the plasma and other membranes.
insoluble fraction
That fraction of cells, prepared by disruptive biochemical methods, that is not soluble in water.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

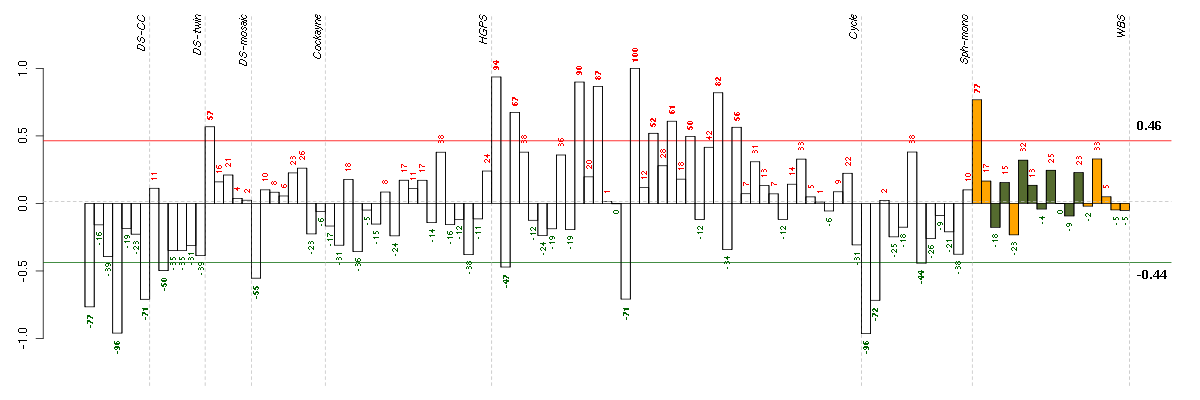
ADORA1adenosine A1 receptor (216220_s_at), score: -0.9 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.95 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.91 F11coagulation factor XI (206610_s_at), score: -0.89 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: -0.97 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: -0.93 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: -0.87 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.96 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.99 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.89 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: -0.88 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: -0.94 RLN1relaxin 1 (211753_s_at), score: -0.88 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: -0.9 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.92 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.88 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -1 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: -0.93
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |