
Under-expression is coded with green,
over-expression with red color.



AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.9 CLCA2chloride channel accessory 2 (217528_at), score: 0.86 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: 1 KRT2keratin 2 (207908_at), score: 0.92 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.98 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.88 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.97 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: 0.93 TAAR2trace amine associated receptor 2 (221394_at), score: 0.89
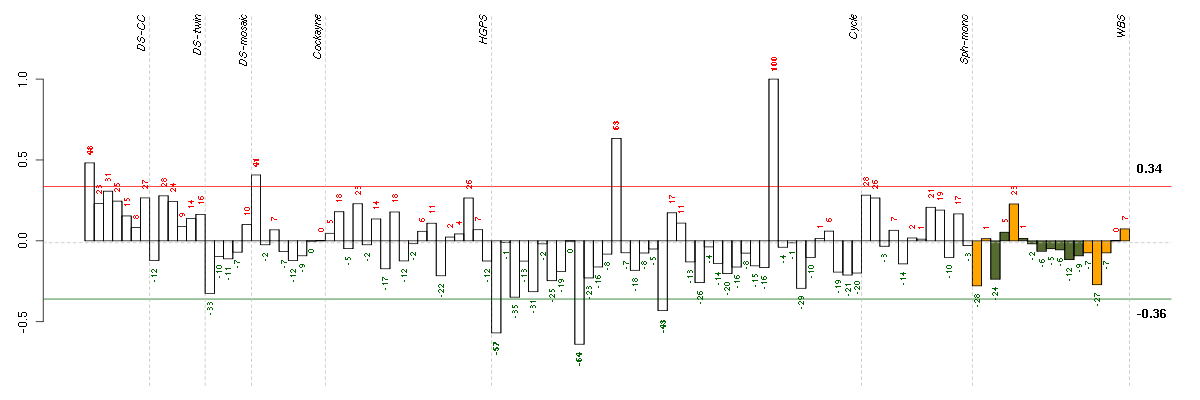
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |