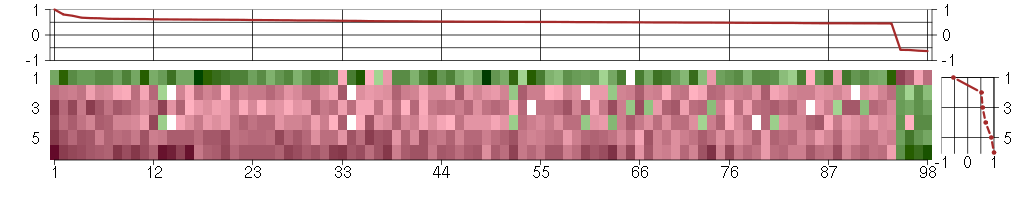

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

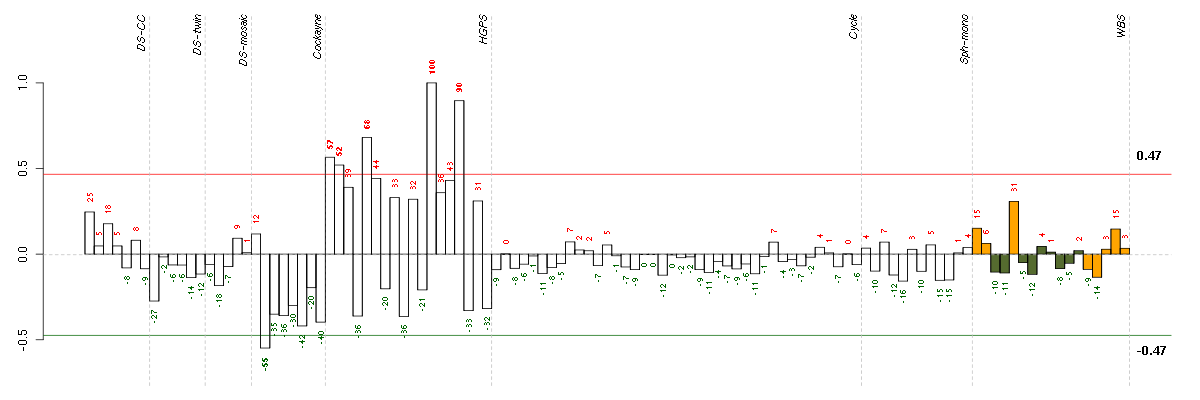
ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.54 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: 0.56 ADORA1adenosine A1 receptor (216220_s_at), score: 0.46 ANTXR1anthrax toxin receptor 1 (220092_s_at), score: 0.59 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.47 AUTS2autism susceptibility candidate 2 (212599_at), score: 0.46 C1orf54chromosome 1 open reading frame 54 (219506_at), score: -0.62 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.46 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.8 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.49 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.51 CDH4cadherin 4, type 1, R-cadherin (retinal) (206866_at), score: 0.53 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 1 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.53 CKBcreatine kinase, brain (200884_at), score: 0.63 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.54 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.49 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.56 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.76 CRATcarnitine acetyltransferase (209522_s_at), score: 0.49 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.54 ELL2elongation factor, RNA polymerase II, 2 (214446_at), score: 0.52 EMX2empty spiracles homeobox 2 (221950_at), score: 0.51 EPB41L3erythrocyte membrane protein band 4.1-like 3 (206710_s_at), score: 0.46 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: 0.6 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.51 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.51 GABRA5gamma-aminobutyric acid (GABA) A receptor, alpha 5 (206456_at), score: 0.56 GLSglutaminase (203159_at), score: 0.5 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.48 GRAMD4GRAM domain containing 4 (212856_at), score: 0.48 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.52 HOXA10homeobox A10 (213150_at), score: 0.51 HOXA11homeobox A11 (213823_at), score: 0.5 HOXA9homeobox A9 (214651_s_at), score: 0.54 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.46 IER3immediate early response 3 (201631_s_at), score: -0.59 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: 0.52 INHBBinhibin, beta B (205258_at), score: 0.63 IRX5iroquois homeobox 5 (210239_at), score: 0.61 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.66 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: 0.49 KRT19keratin 19 (201650_at), score: 0.49 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.6 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.58 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.64 MSX1msh homeobox 1 (205932_s_at), score: 0.57 MT1Mmetallothionein 1M (217546_at), score: 0.51 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.5 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.6 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.61 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.51 NDNnecdin homolog (mouse) (209550_at), score: 0.52 NRG1neuregulin 1 (206343_s_at), score: 0.57 NTF3neurotrophin 3 (206706_at), score: 0.55 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.57 OXTRoxytocin receptor (206825_at), score: 0.47 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.53 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.63 PLAC8placenta-specific 8 (219014_at), score: 0.49 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.46 PODXLpodocalyxin-like (201578_at), score: 0.45 PRPS1phosphoribosyl pyrophosphate synthetase 1 (208447_s_at), score: 0.48 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: 0.58 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.58 PSG6pregnancy specific beta-1-glycoprotein 6 (209738_x_at), score: 0.54 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.59 PTPLBprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (212640_at), score: 0.46 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.66 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.53 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.47 RAB31RAB31, member RAS oncogene family (217763_s_at), score: 0.47 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.5 RUNX3runt-related transcription factor 3 (204198_s_at), score: 0.68 SALL1sal-like 1 (Drosophila) (206893_at), score: 0.5 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.48 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.6 SIRPAsignal-regulatory protein alpha (202897_at), score: 0.57 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.48 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.63 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.46 SLC5A3solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 (213164_at), score: 0.48 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: 0.49 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.58 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.46 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.51 SOCS2suppressor of cytokine signaling 2 (203373_at), score: 0.58 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.5 STMN2stathmin-like 2 (203000_at), score: 0.62 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.61 TBX5T-box 5 (207155_at), score: 0.5 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.63 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.49 TSPAN13tetraspanin 13 (217979_at), score: 0.6 UBL3ubiquitin-like 3 (201535_at), score: 0.51 UNC5Bunc-5 homolog B (C. elegans) (213100_at), score: 0.61 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: 0.52 XYLT1xylosyltransferase I (213725_x_at), score: 0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |