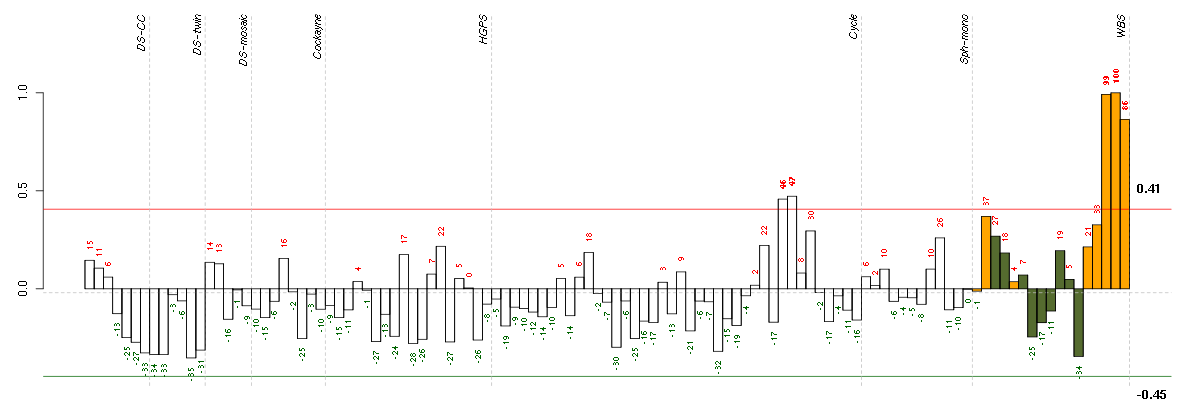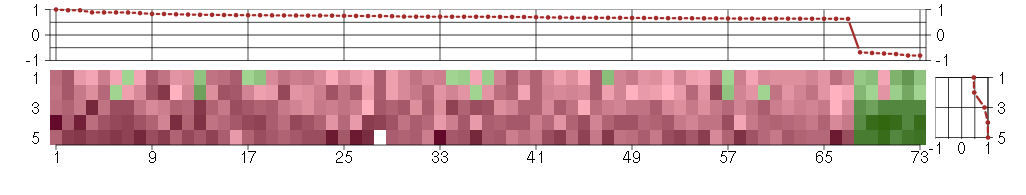

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.64 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.78 AKAP6A kinase (PRKA) anchor protein 6 (205359_at), score: 0.78 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (221589_s_at), score: 0.64 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (209539_at), score: 0.64 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.67 ASPNasporin (219087_at), score: 0.72 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: 0.71 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.76 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.74 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: 0.69 C10orf57chromosome 10 open reading frame 57 (218174_s_at), score: 0.64 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.78 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.74 C4orf30chromosome 4 open reading frame 30 (219717_at), score: 0.65 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: 0.89 CCNG1cyclin G1 (208796_s_at), score: 0.64 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.71 CDH8cadherin 8, type 2 (217574_at), score: 0.71 CRYL1crystallin, lambda 1 (220753_s_at), score: 0.86 CTSOcathepsin O (203758_at), score: 0.78 DBC1deleted in bladder cancer 1 (205818_at), score: -0.75 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.68 F10coagulation factor X (205620_at), score: 0.72 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.68 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.97 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.65 GLRBglycine receptor, beta (205280_at), score: 0.66 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.83 HHLA3HERV-H LTR-associating 3 (220387_s_at), score: 0.72 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.66 HSPB3heat shock 27kDa protein 3 (206375_s_at), score: 0.77 ID1inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (208937_s_at), score: -0.8 IFI35interferon-induced protein 35 (209417_s_at), score: 0.65 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.7 IGF1insulin-like growth factor 1 (somatomedin C) (209541_at), score: -0.68 IL27RAinterleukin 27 receptor, alpha (205926_at), score: 0.79 ITM2Bintegral membrane protein 2B (217732_s_at), score: 0.64 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.67 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: 0.76 LEPRleptin receptor (209894_at), score: 0.72 LEPREL2leprecan-like 2 (204854_at), score: 0.67 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.77 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.76 LMF1lipase maturation factor 1 (219135_s_at), score: 0.67 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: 0.74 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.64 MEGF9multiple EGF-like-domains 9 (212830_at), score: 0.67 MR1major histocompatibility complex, class I-related (207565_s_at), score: 0.81 N4BP2L1NEDD4 binding protein 2-like 1 (213375_s_at), score: 0.72 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.63 NME3non-metastatic cells 3, protein expressed in (204862_s_at), score: 0.88 NRCAMneuronal cell adhesion molecule (204105_s_at), score: -0.73 OCEL1occludin/ELL domain containing 1 (205441_at), score: 0.66 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.76 PAPOLGpoly(A) polymerase gamma (222273_at), score: 0.71 PAPPA2pappalysin 2 (213332_at), score: -0.71 PCYOX1Lprenylcysteine oxidase 1 like (218953_s_at), score: 0.79 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.82 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.75 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: 0.69 SLC46A3solute carrier family 46, member 3 (214719_at), score: 1 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.64 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.81 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.67 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.97 TBC1D12TBC1 domain family, member 12 (221858_at), score: 0.71 TM4SF20transmembrane 4 L six family member 20 (220639_at), score: 0.64 TOMM7translocase of outer mitochondrial membrane 7 homolog (yeast) (201812_s_at), score: 0.72 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.8 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.89 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.89 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: 0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |