

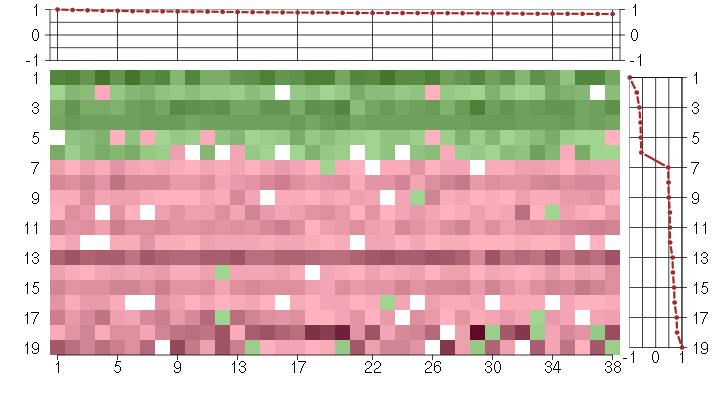

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.88 ADRBK2adrenergic, beta, receptor kinase 2 (204184_s_at), score: 0.83 AGBL2ATP/GTP binding protein-like 2 (220390_at), score: 0.83 ATRNL1attractin-like 1 (213744_at), score: 0.85 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.92 CA1carbonic anhydrase I (205949_at), score: 0.86 CD53CD53 molecule (203416_at), score: 0.98 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: 0.86 CLDN10claudin 10 (205328_at), score: 0.9 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: 0.87 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: 0.89 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.84 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: 0.82 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.92 GRM6glutamate receptor, metabotropic 6 (208035_at), score: 0.88 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.89 KRT2keratin 2 (207908_at), score: 0.93 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.97 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.86 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.93 PEG3paternally expressed 3 (209242_at), score: 0.95 POU4F2POU class 4 homeobox 2 (207725_at), score: 0.91 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.85 PROL1proline rich, lacrimal 1 (208004_at), score: 0.86 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 1 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: 0.85 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.88 SFTPBsurfactant protein B (214354_x_at), score: 0.85 SIRPB1signal-regulatory protein beta 1 (206934_at), score: 0.92 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.83 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: 0.83 STXBP5Lsyntaxin binding protein 5-like (215518_at), score: 0.83 TAAR2trace amine associated receptor 2 (221394_at), score: 0.86 TACR1tachykinin receptor 1 (208048_at), score: 0.95 TAS2R16taste receptor, type 2, member 16 (221444_at), score: 0.89 TP63tumor protein p63 (209863_s_at), score: 0.93 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.87 VPRBPVpr (HIV-1) binding protein (204377_s_at), score: 0.86
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
