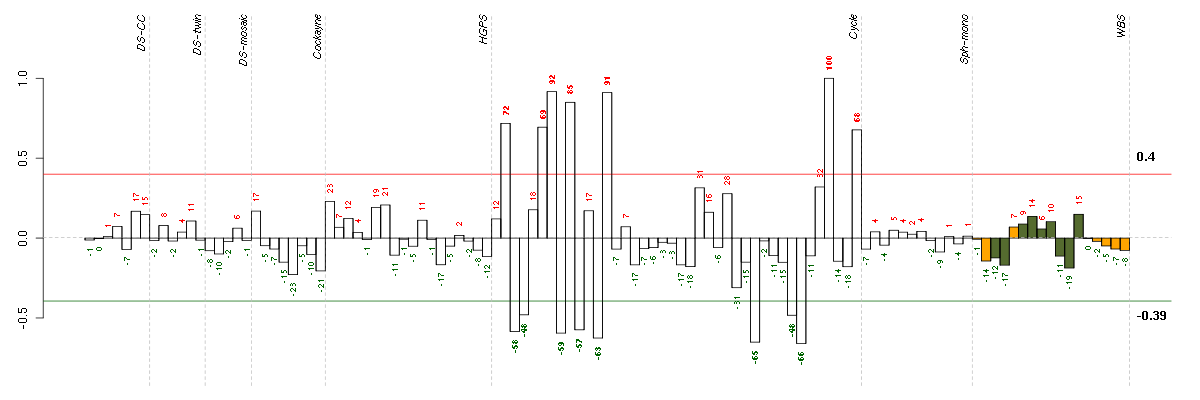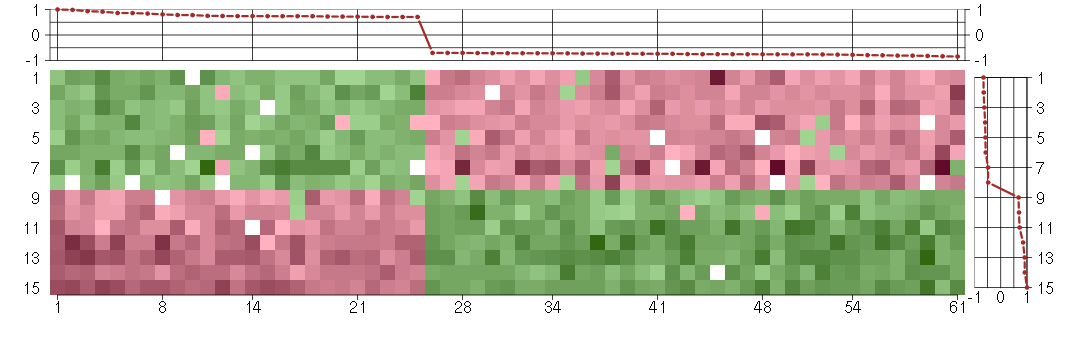

Under-expression is coded with green,
over-expression with red color.



ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.74 ADORA2Badenosine A2b receptor (205891_at), score: -0.73 AIM1absent in melanoma 1 (212543_at), score: -0.75 CALB2calbindin 2 (205428_s_at), score: 0.78 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.75 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: -0.77 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.71 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.82 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.75 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.86 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.74 EPHA4EPH receptor A4 (206114_at), score: 0.71 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.76 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.91 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.81 GMPRguanosine monophosphate reductase (204187_at), score: -0.71 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.72 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.84 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.72 JUPjunction plakoglobin (201015_s_at), score: -0.79 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.98 KIAA1305KIAA1305 (220911_s_at), score: -0.84 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.74 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.86 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.7 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.74 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.74 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.74 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.73 NEFMneurofilament, medium polypeptide (205113_at), score: 1 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.7 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: -0.76 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.84 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.75 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.71 PCDH9protocadherin 9 (219737_s_at), score: 0.93 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.81 PITX1paired-like homeodomain 1 (208502_s_at), score: -0.72 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: -0.73 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.71 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.78 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.76 RNF220ring finger protein 220 (219988_s_at), score: -0.73 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.72 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.72 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.81 SF1splicing factor 1 (208313_s_at), score: -0.7 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.72 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.74 SMAD3SMAD family member 3 (218284_at), score: -0.73 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.75 STIM1stromal interaction molecule 1 (202764_at), score: -0.76 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: -0.76 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.72 TP53tumor protein p53 (201746_at), score: -0.8 TRIM21tripartite motif-containing 21 (204804_at), score: -0.76 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.71 TSKUtsukushin (218245_at), score: -0.71 ULBP2UL16 binding protein 2 (221291_at), score: 0.75 WDR6WD repeat domain 6 (217734_s_at), score: -0.78 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.74
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |