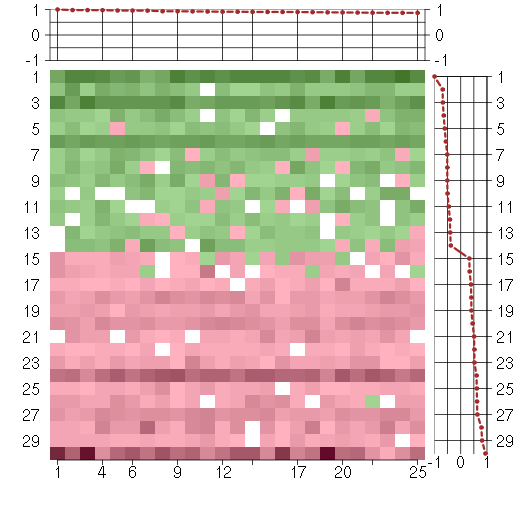

Under-expression is coded with green,
over-expression with red color.



ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.9 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.96 CD53CD53 molecule (203416_at), score: 0.87 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: 0.88 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.9 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: 0.91 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: 0.93 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: 0.96 GK2glycerol kinase 2 (215430_at), score: 0.91 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.87 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.93 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.93 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.89 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.98 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.91 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.92 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.98 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.88 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.87 SFTPBsurfactant protein B (214354_x_at), score: 0.96 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.97 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.9 TBX6T-box 6 (207684_at), score: 0.9 TP63tumor protein p63 (209863_s_at), score: 0.87 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 1
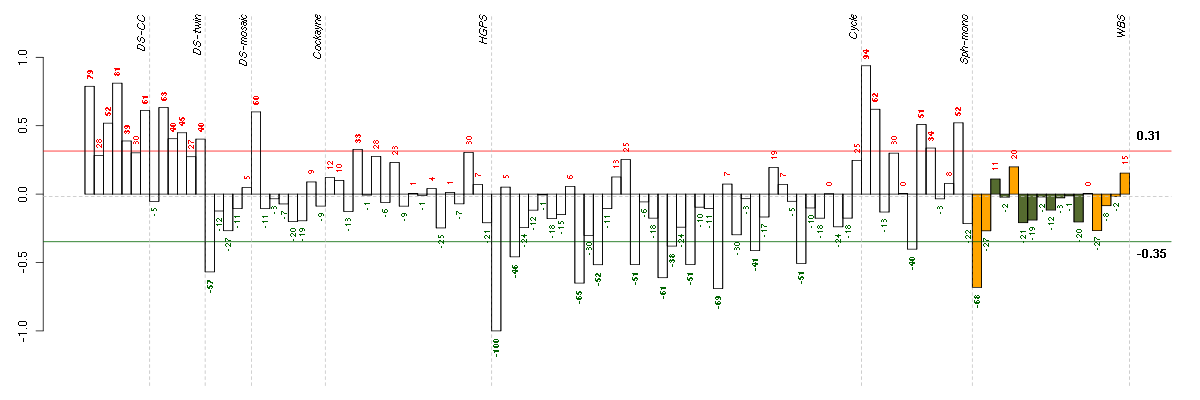
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |