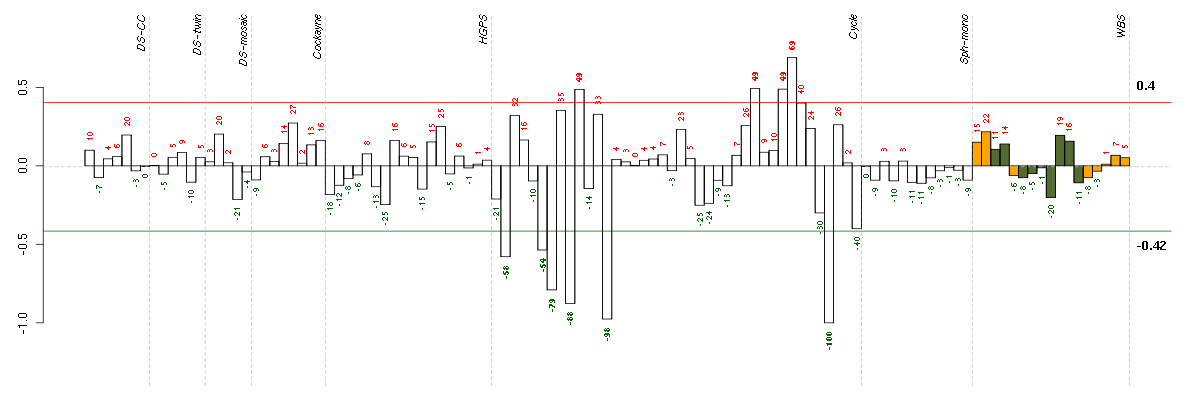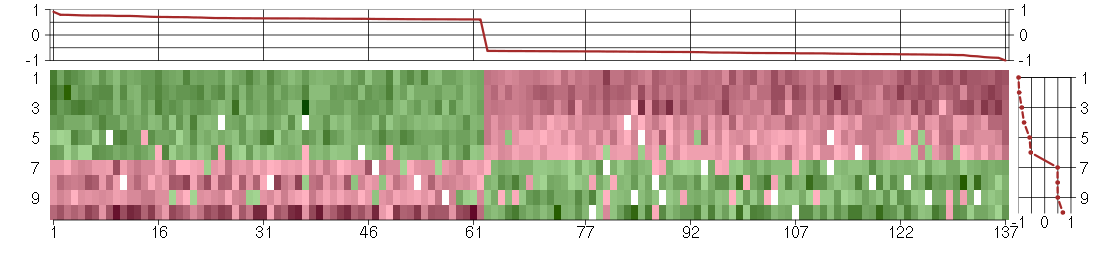

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.

membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.73 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.73 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.73 ADORA2Badenosine A2b receptor (205891_at), score: 0.77 AHI1Abelson helper integration site 1 (221569_at), score: -0.77 AIM1absent in melanoma 1 (212543_at), score: 0.71 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.65 ANXA10annexin A10 (210143_at), score: -0.84 APOL6apolipoprotein L, 6 (219716_at), score: 0.66 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.65 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.64 BIN1bridging integrator 1 (210201_x_at), score: 0.61 BMP6bone morphogenetic protein 6 (206176_at), score: -0.66 BTG2BTG family, member 2 (201236_s_at), score: 0.62 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.67 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.76 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.64 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.75 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.69 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.63 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.71 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.66 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.63 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.91 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.64 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.61 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.64 DEPDC6DEP domain containing 6 (218858_at), score: 0.7 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.7 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.64 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.82 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.7 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.75 EAF2ELL associated factor 2 (219551_at), score: -0.71 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.65 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.77 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.74 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.63 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.65 FNBP1formin binding protein 1 (212288_at), score: 0.64 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.76 GHRgrowth hormone receptor (205498_at), score: -0.65 GKglycerol kinase (207387_s_at), score: -0.76 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.87 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.76 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.67 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.64 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.77 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.74 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.7 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.69 IL33interleukin 33 (209821_at), score: -0.72 IRF1interferon regulatory factor 1 (202531_at), score: 0.62 JUNDjun D proto-oncogene (203751_x_at), score: 0.62 JUPjunction plakoglobin (201015_s_at), score: 0.61 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.89 KIAA1305KIAA1305 (220911_s_at), score: 0.75 KIAA1462KIAA1462 (213316_at), score: 0.69 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.78 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.72 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.62 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.76 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.76 LOC399491LOC399491 protein (214035_x_at), score: 0.7 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.75 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.69 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.67 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.74 MC4Rmelanocortin 4 receptor (221467_at), score: -0.71 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.72 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.63 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.71 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.71 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.66 NCALDneurocalcin delta (211685_s_at), score: -0.66 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.79 NPTX1neuronal pentraxin I (204684_at), score: -0.79 NUP160nucleoporin 160kDa (212709_at), score: -0.66 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.64 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.63 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.78 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.65 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.75 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.71 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.67 PCDH9protocadherin 9 (219737_s_at), score: -0.88 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.66 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.61 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.67 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.66 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.62 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.64 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.62 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.68 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.69 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.76 RNF11ring finger protein 11 (208924_at), score: -0.63 RNF220ring finger protein 220 (219988_s_at), score: 0.64 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.61 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.64 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.65 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.63 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.62 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.63 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.72 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.66 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.79 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.71 SMAD3SMAD family member 3 (218284_at), score: 0.66 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.64 SNNstannin (218032_at), score: 0.65 SSTR1somatostatin receptor 1 (208482_at), score: -0.69 SYNJ1synaptojanin 1 (212990_at), score: -0.64 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.65 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.64 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.65 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.65 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.64 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.62 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.75 TLR3toll-like receptor 3 (206271_at), score: 0.64 TMEM2transmembrane protein 2 (218113_at), score: -0.74 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.63 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.62 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.62 TP53tumor protein p53 (201746_at), score: 0.78 TRIM21tripartite motif-containing 21 (204804_at), score: 0.75 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.65 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.67 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.72 ULBP2UL16 binding protein 2 (221291_at), score: -0.75 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.65 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.63 WDR4WD repeat domain 4 (221632_s_at), score: -0.65 WDR6WD repeat domain 6 (217734_s_at), score: 0.71 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |