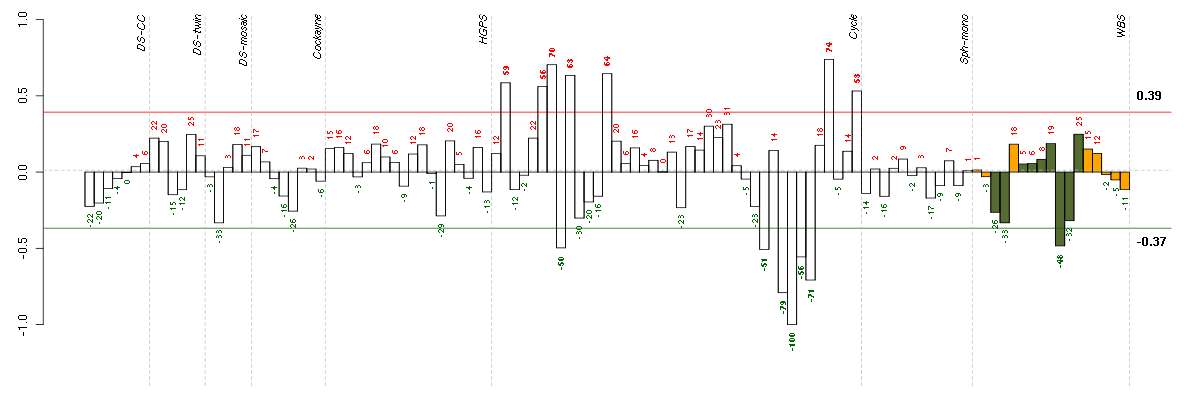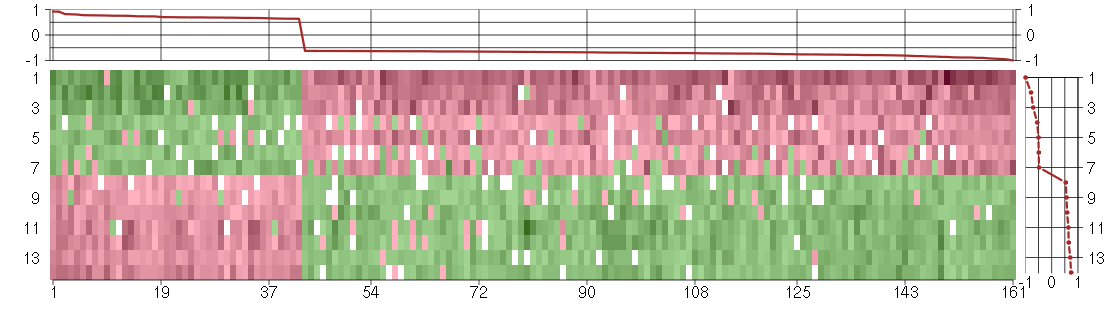

Under-expression is coded with green,
over-expression with red color.

cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
all
This term is the most general term possible

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 2.152e-02 | 1.766 | 8 | 83 | Neuroactive ligand-receptor interaction |
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: -0.73 ABHD3abhydrolase domain containing 3 (213017_at), score: 0.65 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.69 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.86 ADORA2Badenosine A2b receptor (205891_at), score: -0.7 AHI1Abelson helper integration site 1 (221569_at), score: 0.69 AIM1absent in melanoma 1 (212543_at), score: -0.69 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.75 ANGPTL2angiopoietin-like 2 (213001_at), score: -0.93 ANK2ankyrin 2, neuronal (202920_at), score: -0.78 ANXA10annexin A10 (210143_at), score: 0.82 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.62 APOL6apolipoprotein L, 6 (219716_at), score: -0.7 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: -0.65 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (218501_at), score: -0.72 ARL4CADP-ribosylation factor-like 4C (202207_at), score: -0.67 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: -0.8 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: 0.73 BDKRB2bradykinin receptor B2 (205870_at), score: -0.73 BEX4brain expressed, X-linked 4 (215440_s_at), score: -0.65 BIN1bridging integrator 1 (210201_x_at), score: -0.79 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: 0.76 BTG2BTG family, member 2 (201236_s_at), score: -0.83 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: -0.96 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.77 CCNG1cyclin G1 (208796_s_at), score: -0.65 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: -0.82 CLDN11claudin 11 (206908_s_at), score: -0.65 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: -0.77 CRBNcereblon (218142_s_at), score: -0.66 CREB5cAMP responsive element binding protein 5 (205931_s_at), score: -0.63 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.86 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: -0.71 DAPK1death-associated protein kinase 1 (203139_at), score: -0.88 DCLK1doublecortin-like kinase 1 (205399_at), score: -0.88 DDX60DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (218986_s_at), score: -0.64 DENND2ADENN/MADD domain containing 2A (53991_at), score: -0.78 DEPDC6DEP domain containing 6 (218858_at), score: -0.76 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: -0.8 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.7 DOCK10dedicator of cytokinesis 10 (219279_at), score: 0.73 DRAMdamage-regulated autophagy modulator (218627_at), score: -0.65 DTX4deltex homolog 4 (Drosophila) (212611_at), score: -0.69 EFNA5ephrin-A5 (214036_at), score: -0.73 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: -0.84 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: -0.66 FNBP1formin binding protein 1 (212288_at), score: -0.7 FRYfurry homolog (Drosophila) (204072_s_at), score: -0.65 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.66 GALNAC4S-6STB cell RAG associated protein (203066_at), score: -0.73 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: -0.77 GDF15growth differentiation factor 15 (221577_x_at), score: -0.64 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: -0.72 GKglycerol kinase (207387_s_at), score: 0.68 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.75 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: -0.68 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.66 GRK5G protein-coupled receptor kinase 5 (204396_s_at), score: -0.63 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.94 GUCY1B3guanylate cyclase 1, soluble, beta 3 (203817_at), score: -0.7 HERC6hect domain and RLD 6 (219352_at), score: -0.71 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.68 HMGCR3-hydroxy-3-methylglutaryl-Coenzyme A reductase (202540_s_at), score: -0.64 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: -0.73 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.81 IFIH1interferon induced with helicase C domain 1 (219209_at), score: -0.9 IL7Rinterleukin 7 receptor (205798_at), score: 0.67 INSIG1insulin induced gene 1 (201627_s_at), score: -0.65 IRF1interferon regulatory factor 1 (202531_at), score: -0.7 JUNDjun D proto-oncogene (203751_x_at), score: -0.7 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.67 KCNB1potassium voltage-gated channel, Shab-related subfamily, member 1 (211006_s_at), score: -0.76 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: 0.69 KIAA1305KIAA1305 (220911_s_at), score: -0.72 KIAA1462KIAA1462 (213316_at), score: -0.69 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.69 LDLRlow density lipoprotein receptor (202068_s_at), score: -0.79 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: 0.64 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.69 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.74 LOC399491LOC399491 protein (214035_x_at), score: -0.65 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.7 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: -0.75 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: 0.64 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: -0.67 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -1 MBPmyelin basic protein (210136_at), score: -0.68 MC4Rmelanocortin 4 receptor (221467_at), score: 0.8 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: 0.73 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.68 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: 0.78 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: 0.81 MSX2msh homeobox 2 (210319_x_at), score: -0.76 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: -0.73 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: -0.92 NEFMneurofilament, medium polypeptide (205113_at), score: 0.92 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: 0.73 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.72 NPTX1neuronal pentraxin I (204684_at), score: 0.77 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: -0.63 NTMneurotrimin (222020_s_at), score: 0.69 NUP160nucleoporin 160kDa (212709_at), score: 0.64 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: -0.84 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.76 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.88 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.63 PCDH9protocadherin 9 (219737_s_at), score: 0.75 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.65 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: 0.64 PDE4Aphosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) (204735_at), score: -0.64 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.63 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: -0.64 PDSS1prenyl (decaprenyl) diphosphate synthase, subunit 1 (220865_s_at), score: 0.68 PLEKHA5pleckstrin homology domain containing, family A member 5 (220952_s_at), score: -0.67 PMP22peripheral myelin protein 22 (210139_s_at), score: -0.63 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: -0.66 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: 0.77 PPLperiplakin (203407_at), score: -0.87 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: 0.75 PRR5proline rich 5 (renal) (47069_at), score: -0.68 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.68 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: -0.68 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.83 RCAN2regulator of calcineurin 2 (203498_at), score: -0.73 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: -0.63 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: -0.67 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: -0.79 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: -0.63 RPL13Aribosomal protein L13a (200715_x_at), score: -0.65 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: 0.7 SECTM1secreted and transmembrane 1 (213716_s_at), score: -0.79 SELENBP1selenium binding protein 1 (214433_s_at), score: -0.72 SESN1sestrin 1 (218346_s_at), score: -0.64 SIX2SIX homeobox 2 (206510_at), score: -0.75 SLC1A3solute carrier family 1 (glial high affinity glutamate transporter), member 3 (202800_at), score: -0.63 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.77 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: -0.63 SNNstannin (218032_at), score: -0.66 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: -0.63 SQLEsqualene epoxidase (213562_s_at), score: -0.64 SSTR1somatostatin receptor 1 (208482_at), score: 0.67 SYNPOsynaptopodin (202796_at), score: -0.76 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: -0.71 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.9 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: -0.67 TLR3toll-like receptor 3 (206271_at), score: -0.76 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: 0.67 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.63 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: -0.79 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: -0.63 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: -0.69 TNS3tensin 3 (217853_at), score: -0.66 TP53tumor protein p53 (201746_at), score: -0.78 TRIM21tripartite motif-containing 21 (204804_at), score: -0.69 TSPAN13tetraspanin 13 (217979_at), score: 0.69 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.65 ULBP2UL16 binding protein 2 (221291_at), score: 0.92 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.63 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: -0.71 XAF1XIAP associated factor 1 (206133_at), score: -0.66 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |