
Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome membrane
The lipid bilayer surrounding a melanosome.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
chitosome
An intracellular membrane-bounded particle found in fungi and containing chitin synthase; it synthesizes chitin microfibrils. Chitin synthase activity exists in chitosomes and they are proposed to act as a reservoir for regulated transport of chitin synthase enzymes to the division septum.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
melanosome membrane
The lipid bilayer surrounding a melanosome.
melanosome membrane
The lipid bilayer surrounding a melanosome.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
UDP-glycosyltransferase activity
Catalysis of the transfer of a glycosyl group from a UDP-sugar to a small hydrophobic molecule.
glucuronosyltransferase activity
Catalysis of the reaction: UDP-glucuronate + acceptor = UDP + acceptor beta-D-glucuronoside.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring glycosyl groups
Catalysis of the transfer of a glycosyl group from one compound (donor) to another (acceptor).
transferase activity, transferring hexosyl groups
Catalysis of the transfer of a hexosyl group from one compound (donor) to another (acceptor).
all
This term is the most general term possible
glucuronosyltransferase activity
Catalysis of the reaction: UDP-glucuronate + acceptor = UDP + acceptor beta-D-glucuronoside.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00500 | 9.080e-04 | 0.2817 | 5 | 24 | Starch and sucrose metabolism |
| 00860 | 9.080e-04 | 0.2817 | 5 | 24 | Porphyrin and chlorophyll metabolism |
| 00040 | 1.432e-03 | 0.1526 | 4 | 13 | Pentose and glucuronate interconversions |
| 00150 | 8.054e-03 | 0.2465 | 4 | 21 | Androgen and estrogen metabolism |
| 00830 | 1.101e-02 | 0.27 | 4 | 23 | Retinol metabolism |
| 00983 | 2.191e-02 | 0.3404 | 4 | 29 | Drug metabolism - other enzymes |
| 00982 | 4.165e-02 | 0.4226 | 4 | 36 | Drug metabolism - cytochrome P450 |
| 00980 | 4.517e-02 | 0.4343 | 4 | 37 | Metabolism of xenobiotics by cytochrome P450 |
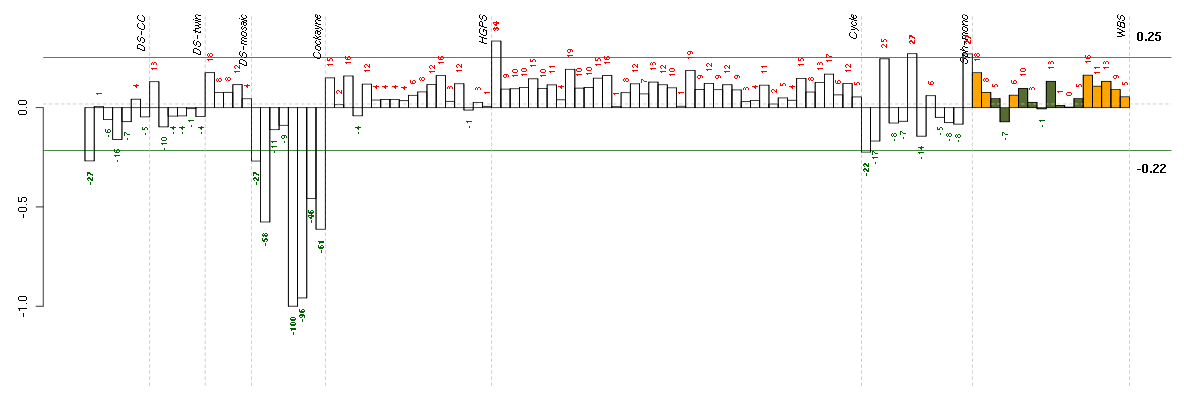
A2Malpha-2-macroglobulin (217757_at), score: -0.59 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -1 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: -0.68 AIM2absent in melanoma 2 (206513_at), score: -0.66 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.49 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (201612_at), score: 0.64 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.54 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: 0.61 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.49 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.47 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.61 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.68 CA3carbonic anhydrase III, muscle specific (204865_at), score: -0.72 CD24CD24 molecule (209771_x_at), score: -0.5 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: -0.47 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.62 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.67 CRTAPcartilage associated protein (201380_at), score: 0.81 CXorf21chromosome X open reading frame 21 (220252_x_at), score: -0.47 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.68 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: -0.73 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.5 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: -0.54 EPHB2EPH receptor B2 (209589_s_at), score: -0.52 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.57 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.64 FBN2fibrillin 2 (203184_at), score: 0.64 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.73 FZD3frizzled homolog 3 (Drosophila) (219683_at), score: -0.56 GAGE4G antigen 4 (207086_x_at), score: -0.97 GAGE6G antigen 6 (208155_x_at), score: -0.89 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: 0.61 GATA3GATA binding protein 3 (209604_s_at), score: -0.92 HERC5hect domain and RLD 5 (219863_at), score: -0.76 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.78 HLA-DPA1major histocompatibility complex, class II, DP alpha 1 (211990_at), score: -0.53 HRASLSHRAS-like suppressor (219983_at), score: -0.58 HTR1F5-hydroxytryptamine (serotonin) receptor 1F (221458_at), score: -0.5 KIAA0495KIAA0495 (213340_s_at), score: 0.62 KIF13Bkinesin family member 13B (202962_at), score: -0.47 KIFC1kinesin family member C1 (209680_s_at), score: -0.47 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.68 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.49 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.51 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.53 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.63 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.53 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.7 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.69 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.49 MRC2mannose receptor, C type 2 (37408_at), score: 0.65 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: -0.47 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: 0.64 MYO1Cmyosin IC (214656_x_at), score: 0.72 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.67 NEFLneurofilament, light polypeptide (221805_at), score: -0.63 NMUneuromedin U (206023_at), score: -0.7 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.49 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.67 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.99 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.64 PDE10Aphosphodiesterase 10A (205501_at), score: -0.66 PDE8Bphosphodiesterase 8B (213228_at), score: -0.48 PGK2phosphoglycerate kinase 2 (217009_at), score: -0.49 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.58 PIWIL1piwi-like 1 (Drosophila) (214868_at), score: -0.52 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.48 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.63 PRKCHprotein kinase C, eta (218764_at), score: -0.51 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.68 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.72 REEP1receptor accessory protein 1 (204364_s_at), score: -0.57 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.49 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.69 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.63 ST3GAL6ST3 beta-galactoside alpha-2,3-sialyltransferase 6 (210942_s_at), score: -0.47 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.57 TAC1tachykinin, precursor 1 (206552_s_at), score: -0.61 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.5 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.55 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.88 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.61 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: -0.49 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.65 TMEM176Btransmembrane protein 176B (220532_s_at), score: -0.51 TMEM47transmembrane protein 47 (209656_s_at), score: 0.61 TMOD1tropomodulin 1 (203661_s_at), score: -0.6 TNS1tensin 1 (221748_s_at), score: 0.65 TOB1transducer of ERBB2, 1 (202704_at), score: 0.61 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.6 UBL3ubiquitin-like 3 (201535_at), score: 0.69 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.96 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.96 UGT1A8UDP glucuronosyltransferase 1 family, polypeptide A8 (221304_at), score: -0.57 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.94 UROSuroporphyrinogen III synthase (203031_s_at), score: 0.61 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.65 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.55 WT1Wilms tumor 1 (206067_s_at), score: -0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |