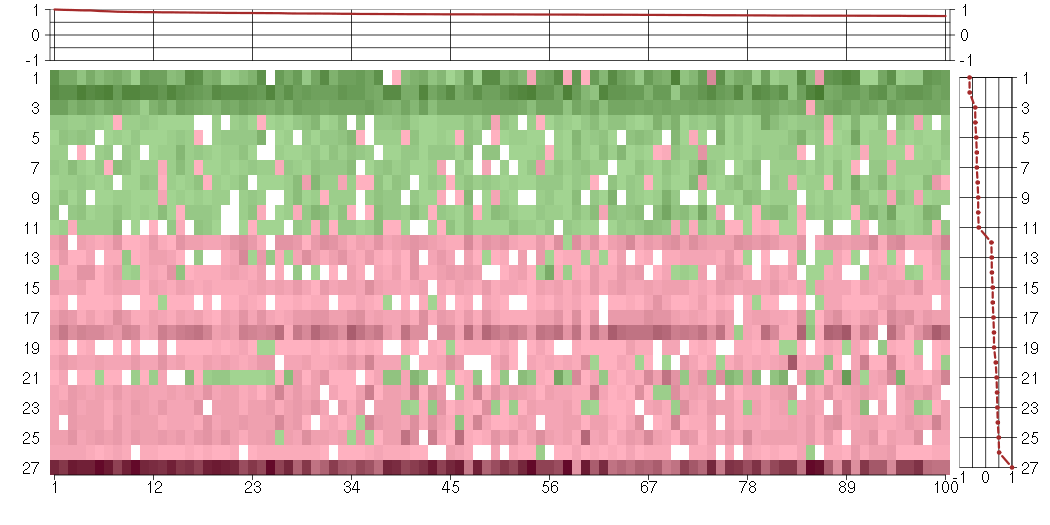

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
acrosome
A structure in the head of a spermatozoon that contains acid hydrolases, and is concerned with the breakdown of the outer membrane of the ovum during fertilization. It lies just beneath the plasma membrane and is derived from the lysosome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00830 | 3.040e-03 | 0.1856 | 4 | 23 | Retinol metabolism |
| 00982 | 1.383e-02 | 0.2905 | 4 | 36 | Drug metabolism - cytochrome P450 |
| 00980 | 1.489e-02 | 0.2986 | 4 | 37 | Metabolism of xenobiotics by cytochrome P450 |
ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.76 ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.88 ADORA1adenosine A1 receptor (216220_s_at), score: 0.75 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.8 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: 0.91 ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.87 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.8 ATP5OATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (216954_x_at), score: 0.76 BCL9B-cell CLL/lymphoma 9 (204129_at), score: 0.75 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.81 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.79 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.74 CDR1cerebellar degeneration-related protein 1, 34kDa (207276_at), score: 0.81 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: 0.76 CEP27centrosomal protein 27kDa (220071_x_at), score: 0.8 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: 0.93 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: 0.79 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.81 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: 0.84 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.76 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.9 DAZ1deleted in azoospermia 1 (216922_x_at), score: 0.82 DAZ3deleted in azoospermia 3 (208281_x_at), score: 0.83 DAZ4deleted in azoospermia 4 (216351_x_at), score: 0.86 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.76 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: 0.89 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.82 EPAGearly lymphoid activation protein (217050_at), score: 0.83 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: 0.79 FBXO41F-box protein 41 (44040_at), score: 0.86 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 0.9 FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.88 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: 0.84 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.94 GK2glycerol kinase 2 (215430_at), score: 0.81 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.77 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.79 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.82 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.85 IFNA16interferon, alpha 16 (208448_x_at), score: 0.78 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.79 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.79 LOC100129624hypothetical LOC100129624 (210748_at), score: 0.81 LOC100130233similar to NPM1 protein (216387_x_at), score: 0.81 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.75 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.77 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.77 LOC91316glucuronidase, beta/ immunoglobulin lambda-like polypeptide 1 pseudogene (215816_at), score: 0.74 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.77 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.81 MEFVMediterranean fever (208262_x_at), score: 0.8 METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.91 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.96 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.77 MKRN1makorin ring finger protein 1 (208082_x_at), score: 0.88 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.82 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.81 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.75 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.75 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.9 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.78 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.82 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 1 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.78 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.79 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: 0.97 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.81 RPL35Aribosomal protein L35a (215208_x_at), score: 0.86 RPS10P3ribosomal protein S10 pseudogene 3 (217336_at), score: 0.77 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: 0.76 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.81 SCN11Asodium channel, voltage-gated, type XI, alpha subunit (220791_x_at), score: 0.77 SFTPBsurfactant protein B (214354_x_at), score: 0.84 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.87 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.8 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.76 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.76 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.87 SPNsialophorin (206057_x_at), score: 0.8 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 0.98 SYCP1synaptonemal complex protein 1 (206740_x_at), score: 0.84 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 0.86 TP53TG3TP53 target 3 (220167_s_at), score: 0.8 TRIM36tripartite motif-containing 36 (219736_at), score: 0.81 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.76 UBQLN4ubiquilin 4 (222252_x_at), score: 0.82 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.99 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.84 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: 0.78 VPRBPVpr (HIV-1) binding protein (204377_s_at), score: 0.76 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: 0.86 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.76 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.81 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.87 ZNF117zinc finger protein 117 (207605_x_at), score: 0.75 ZNF160zinc finger protein 160 (214715_x_at), score: 0.75 ZNF440zinc finger protein 440 (215892_at), score: 0.8 ZNF492zinc finger protein 492 (215532_x_at), score: 0.79 ZNF639zinc finger protein 639 (218413_s_at), score: 0.82 ZNF816Azinc finger protein 816A (217541_x_at), score: 0.89
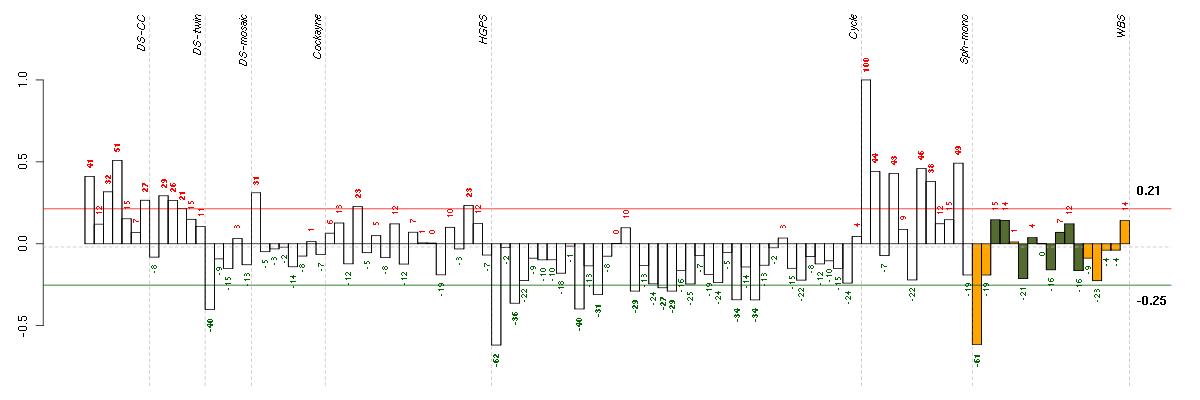
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |