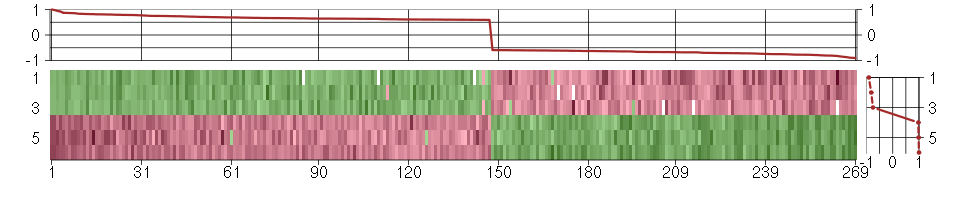

Under-expression is coded with green,
over-expression with red color.

angiogenesis
Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels.
blood vessel development
The process whose specific outcome is the progression of the blood vessel over time, from its formation to the mature structure. The blood vessel is the vasculature carrying blood.
vasculature development
The process whose specific outcome is the progression of the vasculature over time, from its formation to the mature structure.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signal transduction
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
locomotion
Self-propelled movement of a cell or organism from one location to another.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
blood vessel morphogenesis
The process by which the anatomical structures of blood vessels are generated and organized. Morphogenesis pertains to the creation of form. The blood vessel is the vasculature carrying blood.
anatomical structure formation
The process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
anatomical structure formation
The process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
angiogenesis
Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels.
blood vessel morphogenesis
The process by which the anatomical structures of blood vessels are generated and organized. Morphogenesis pertains to the creation of form. The blood vessel is the vasculature carrying blood.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intermediate filament cytoskeleton
Cytoskeletal structure made from intermediate filaments, typically organized in the cytosol as an extended system that stretches from the nuclear envelope to the plasma membrane. Some intermediate filaments run parallel to the cell surface, while others traverse the cytosol; together they form an internal framework that helps support the shape and resilience of the cell.
neurofilament cytoskeleton
Intermediate filament cytoskeletal structure that is made up of neurofilaments. Neurofilaments are specialized intermediate filaments found in neurons.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
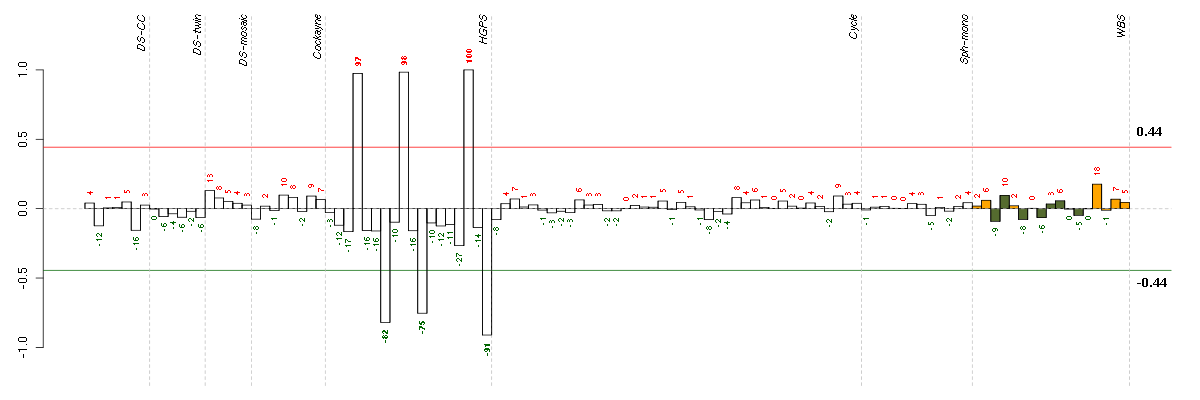
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: -0.74 ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: 0.81 ACANaggrecan (205679_x_at), score: 0.64 ACTG2actin, gamma 2, smooth muscle, enteric (202274_at), score: 0.65 ADAM23ADAM metallopeptidase domain 23 (206046_at), score: 0.65 ADAMTSL3ADAMTS-like 3 (213974_at), score: -0.71 AGTR1angiotensin II receptor, type 1 (205357_s_at), score: 0.71 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.68 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.63 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: 0.78 ANKRD6ankyrin repeat domain 6 (204671_s_at), score: 0.79 ANO3anoctamin 3 (215241_at), score: 0.71 ANXA3annexin A3 (209369_at), score: -0.73 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.61 APOLD1apolipoprotein L domain containing 1 (221031_s_at), score: 0.65 AQP1aquaporin 1 (Colton blood group) (209047_at), score: 0.7 ARFIP2ADP-ribosylation factor interacting protein 2 (202109_at), score: 0.61 ARHGAP17Rho GTPase activating protein 17 (218076_s_at), score: 0.6 ARHGAP29Rho GTPase activating protein 29 (203910_at), score: -0.81 ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (221656_s_at), score: 0.63 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.62 ATF3activating transcription factor 3 (202672_s_at), score: 0.77 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (203304_at), score: 0.62 BCL11AB-cell CLL/lymphoma 11A (zinc finger protein) (219497_s_at), score: 0.61 BMP6bone morphogenetic protein 6 (206176_at), score: -0.61 BNC2basonuclin 2 (220272_at), score: -0.8 BTG3BTG family, member 3 (213134_x_at), score: -0.6 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: 0.59 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.66 C3complement component 3 (217767_at), score: -0.62 C8orf84chromosome 8 open reading frame 84 (214725_at), score: -0.71 C9orf95chromosome 9 open reading frame 95 (219147_s_at), score: 0.64 CARHSP1calcium regulated heat stable protein 1, 24kDa (218384_at), score: -0.75 CCDC68coiled-coil domain containing 68 (220180_at), score: -0.63 CCDC81coiled-coil domain containing 81 (220389_at), score: 0.75 CCDC92coiled-coil domain containing 92 (218175_at), score: 0.7 CCND2cyclin D2 (200953_s_at), score: 0.66 CD200CD200 molecule (209583_s_at), score: -0.75 CD34CD34 molecule (209543_s_at), score: -0.67 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.9 CDK5R1cyclin-dependent kinase 5, regulatory subunit 1 (p35) (204995_at), score: 0.64 CENPQcentromere protein Q (219294_at), score: -0.67 CFHcomplement factor H (213800_at), score: -0.72 CFHR1complement factor H-related 1 (215388_s_at), score: -0.72 CHRDL1chordin-like 1 (209763_at), score: -0.75 CLDN1claudin 1 (218182_s_at), score: -0.6 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.68 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.59 CORINcorin, serine peptidase (220356_at), score: 0.83 CRLF1cytokine receptor-like factor 1 (206315_at), score: -0.61 CRYABcrystallin, alpha B (209283_at), score: 0.6 CRYBB2P1crystallin, beta B2 pseudogene 1 (222048_at), score: 0.61 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.77 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: -0.74 CXCL12chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) (209687_at), score: 0.61 CYB5Acytochrome b5 type A (microsomal) (215726_s_at), score: -0.73 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.8 CYTIPcytohesin 1 interacting protein (209606_at), score: 0.66 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.76 DLEU1deleted in lymphocytic leukemia 1 (non-protein coding) (205677_s_at), score: -0.67 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.6 DNAJB5DnaJ (Hsp40) homolog, subfamily B, member 5 (212817_at), score: 0.61 DPYSL3dihydropyrimidinase-like 3 (201430_s_at), score: -0.82 DSG2desmoglein 2 (217901_at), score: -0.81 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.63 EDN1endothelin 1 (218995_s_at), score: -0.7 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.83 EHD3EH-domain containing 3 (218935_at), score: 0.63 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: -0.61 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.67 EN1engrailed homeobox 1 (220559_at), score: 0.79 ENPEPglutamyl aminopeptidase (aminopeptidase A) (204844_at), score: 0.77 EREGepiregulin (205767_at), score: 0.69 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (213541_s_at), score: -0.67 FBLN1fibulin 1 (202995_s_at), score: -0.7 FBLN2fibulin 2 (203886_s_at), score: 0.8 FGL2fibrinogen-like 2 (204834_at), score: -0.82 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.6 FKBP1BFK506 binding protein 1B, 12.6 kDa (206857_s_at), score: -0.6 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.73 FOXO1forkhead box O1 (202724_s_at), score: -0.72 FUT8fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (203988_s_at), score: -0.77 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: -0.66 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.67 GCAgrancalcin, EF-hand calcium binding protein (203765_at), score: 0.59 GPR116G protein-coupled receptor 116 (212950_at), score: 1 GPR126G protein-coupled receptor 126 (213094_at), score: -0.75 GPR4G protein-coupled receptor 4 (206236_at), score: -0.77 GPRC5AG protein-coupled receptor, family C, group 5, member A (203108_at), score: -0.67 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.71 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.64 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.64 GSTT2glutathione S-transferase theta 2 (205439_at), score: -0.59 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: -0.6 HCLS1hematopoietic cell-specific Lyn substrate 1 (202957_at), score: 0.77 HNMThistamine N-methyltransferase (204112_s_at), score: 0.7 HOXB7homeobox B7 (204779_s_at), score: -0.71 HRH1histamine receptor H1 (205579_at), score: -0.76 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: 0.79 ID1inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (208937_s_at), score: -0.7 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.62 IFI27interferon, alpha-inducible protein 27 (202411_at), score: -0.71 IL12Ainterleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) (207160_at), score: 0.61 IL18interleukin 18 (interferon-gamma-inducing factor) (206295_at), score: -0.68 IL1Ainterleukin 1, alpha (210118_s_at), score: -0.65 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.69 IRF1interferon regulatory factor 1 (202531_at), score: 0.68 ITGA7integrin, alpha 7 (216331_at), score: 0.75 ITGA8integrin, alpha 8 (214265_at), score: 0.94 ITM2Aintegral membrane protein 2A (202746_at), score: -0.64 IVNS1ABPinfluenza virus NS1A binding protein (206245_s_at), score: -0.64 JAM3junctional adhesion molecule 3 (212813_at), score: -0.59 KANK2KN motif and ankyrin repeat domains 2 (218418_s_at), score: 0.64 KCND2potassium voltage-gated channel, Shal-related subfamily, member 2 (207103_at), score: 0.8 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.68 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: -0.86 KIAA0247KIAA0247 (202181_at), score: -0.66 KIRRELkin of IRRE like (Drosophila) (220825_s_at), score: 0.62 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: -0.78 KRT7keratin 7 (209016_s_at), score: -0.63 LAMA4laminin, alpha 4 (202202_s_at), score: -0.9 LAMC2laminin, gamma 2 (202267_at), score: -0.75 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.6 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: -0.6 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (213880_at), score: 0.86 LIMCH1LIM and calponin homology domains 1 (212327_at), score: -0.72 LIPGlipase, endothelial (219181_at), score: -0.78 LMO4LIM domain only 4 (209205_s_at), score: -0.61 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: 0.67 LPPR4plasticity related gene 1 (213496_at), score: 0.81 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.69 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.64 MAPRE2microtubule-associated protein, RP/EB family, member 2 (202501_at), score: 0.66 MATN3matrilin 3 (206091_at), score: -0.67 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.62 MCCC1methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) (218440_at), score: 0.61 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: -0.63 MFAP2microfibrillar-associated protein 2 (203417_at), score: 0.62 MFAP5microfibrillar associated protein 5 (213764_s_at), score: -0.62 MFGE8milk fat globule-EGF factor 8 protein (210605_s_at), score: 0.67 MMDmonocyte to macrophage differentiation-associated (203414_at), score: -0.67 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: -0.77 MPDZmultiple PDZ domain protein (213306_at), score: -0.63 MRASmuscle RAS oncogene homolog (206538_at), score: -0.65 MYH10myosin, heavy chain 10, non-muscle (212372_at), score: -0.89 MYLKmyosin light chain kinase (202555_s_at), score: 0.74 MYO1Emyosin IE (203072_at), score: 0.64 NAAAN-acylethanolamine acid amidase (214765_s_at), score: 0.69 NAGLUN-acetylglucosaminidase, alpha- (204360_s_at), score: 0.64 NAP1L3nucleosome assembly protein 1-like 3 (204749_at), score: -0.71 NBEAneurobeachin (221207_s_at), score: 0.64 NEFMneurofilament, medium polypeptide (205113_at), score: 0.81 NESnestin (218678_at), score: 0.67 NFIBnuclear factor I/B (209289_at), score: -0.74 NMUneuromedin U (206023_at), score: 0.66 NOTCH1Notch homolog 1, translocation-associated (Drosophila) (218902_at), score: 0.64 NOX4NADPH oxidase 4 (219773_at), score: -0.81 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.63 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.61 NQO2NAD(P)H dehydrogenase, quinone 2 (203814_s_at), score: -0.67 NRP2neuropilin 2 (214632_at), score: -0.63 NRXN3neurexin 3 (205795_at), score: 0.59 NUDT2nudix (nucleoside diphosphate linked moiety X)-type motif 2 (218609_s_at), score: -0.68 OSBPL1Aoxysterol binding protein-like 1A (209485_s_at), score: 0.69 PARP3poly (ADP-ribose) polymerase family, member 3 (209940_at), score: 0.82 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: 0.66 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.61 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.62 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.63 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.78 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.59 PELOpelota homolog (Drosophila) (218472_s_at), score: 0.61 PFTK1PFTAIRE protein kinase 1 (204604_at), score: -0.64 PHF11PHD finger protein 11 (221816_s_at), score: 0.7 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.62 PIK3R1phosphoinositide-3-kinase, regulatory subunit 1 (alpha) (212240_s_at), score: 0.66 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.67 PKP4plakophilin 4 (201928_at), score: -0.6 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.59 PLCE1phospholipase C, epsilon 1 (205112_at), score: -0.68 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.75 PLXDC1plexin domain containing 1 (219700_at), score: 0.65 PLXNA2plexin A2 (213030_s_at), score: -0.84 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.6 PORCNporcupine homolog (Drosophila) (219483_s_at), score: 0.82 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: 0.72 PPM1Hprotein phosphatase 1H (PP2C domain containing) (212686_at), score: -0.61 PPP1R13Lprotein phosphatase 1, regulatory (inhibitor) subunit 13 like (218849_s_at), score: -0.78 PRNPprion protein (201300_s_at), score: -0.64 PROCRprotein C receptor, endothelial (EPCR) (203650_at), score: 0.65 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: 0.75 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.75 PRSS3protease, serine, 3 (207463_x_at), score: 0.85 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.6 PTGESprostaglandin E synthase (210367_s_at), score: -0.66 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.98 PYGLphosphorylase, glycogen, liver (202990_at), score: -0.6 RAC2ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) (213603_s_at), score: 0.68 RAD51CRAD51 homolog C (S. cerevisiae) (209849_s_at), score: -0.59 RGS14regulator of G-protein signaling 14 (38290_at), score: 0.6 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: -0.76 RPA1replication protein A1, 70kDa (201529_s_at), score: -0.61 RPP25ribonuclease P/MRP 25kDa subunit (219143_s_at), score: 0.87 RTN1reticulon 1 (203485_at), score: 0.92 S100A3S100 calcium binding protein A3 (206027_at), score: 0.63 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.79 SCRN1secernin 1 (201462_at), score: 0.65 SEPT6septin 6 (212414_s_at), score: -0.7 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.62 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: -0.66 SETD8SET domain containing (lysine methyltransferase) 8 (220200_s_at), score: -0.59 SFRP4secreted frizzled-related protein 4 (204051_s_at), score: -0.63 SIGLEC15sialic acid binding Ig-like lectin 15 (215856_at), score: 0.74 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.86 SIX2SIX homeobox 2 (206510_at), score: 0.71 SLC17A9solute carrier family 17, member 9 (219559_at), score: 0.74 SLC22A17solute carrier family 22, member 17 (218675_at), score: -0.61 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (57588_at), score: 0.87 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: -0.68 SLC7A5solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 (201195_s_at), score: -0.77 SMAD6SMAD family member 6 (207069_s_at), score: -0.7 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (214708_at), score: 0.61 SORBS2sorbin and SH3 domain containing 2 (204288_s_at), score: 0.71 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: -0.78 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: 0.65 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.78 STARD7StAR-related lipid transfer (START) domain containing 7 (200028_s_at), score: 0.67 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.64 STBD1starch binding domain 1 (203986_at), score: 0.72 STXBP2syntaxin binding protein 2 (209367_at), score: 0.64 STYK1serine/threonine/tyrosine kinase 1 (221696_s_at), score: 0.67 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.6 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.61 SWAP70SWAP-70 protein (209307_at), score: -0.64 SYNMsynemin, intermediate filament protein (212730_at), score: 0.72 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.61 TBXA2Rthromboxane A2 receptor (336_at), score: 0.81 TEAD3TEA domain family member 3 (209454_s_at), score: 0.75 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: -0.62 THY1Thy-1 cell surface antigen (208850_s_at), score: 0.63 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: 0.73 TLR4toll-like receptor 4 (221060_s_at), score: -0.63 TMEM35transmembrane protein 35 (219685_at), score: 0.64 TMEM80transmembrane protein 80 (65630_at), score: 0.6 TMEM97transmembrane protein 97 (212279_at), score: -0.64 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: -0.6 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.59 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.68 TRIM34tripartite motif-containing 34 (221044_s_at), score: 0.74 TRPV2transient receptor potential cation channel, subfamily V, member 2 (219282_s_at), score: 0.73 TRY6trypsinogen C (215395_x_at), score: 0.82 TSPAN12tetraspanin 12 (219274_at), score: 0.68 TSPYL5TSPY-like 5 (213122_at), score: -0.79 TTLL7tubulin tyrosine ligase-like family, member 7 (219882_at), score: 0.6 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.64 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (214755_at), score: 0.75 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.6 ULBP1UL16 binding protein 1 (221323_at), score: 0.61 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: 0.83 WBP2WW domain binding protein 2 (209117_at), score: 0.6 WNT10Bwingless-type MMTV integration site family, member 10B (206213_at), score: 0.67 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.66 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: 0.7 ZFHX4zinc finger homeobox 4 (219779_at), score: -0.87 ZFP36zinc finger protein 36, C3H type, homolog (mouse) (201531_at), score: 0.69 ZFPM2zinc finger protein, multitype 2 (219778_at), score: -0.71 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.6 ZNF323zinc finger protein 323 (222016_s_at), score: -0.64 ZNF365zinc finger protein 365 (206448_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |