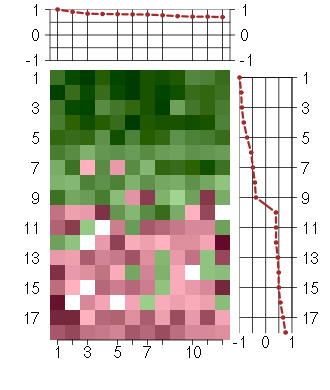

Under-expression is coded with green,
over-expression with red color.



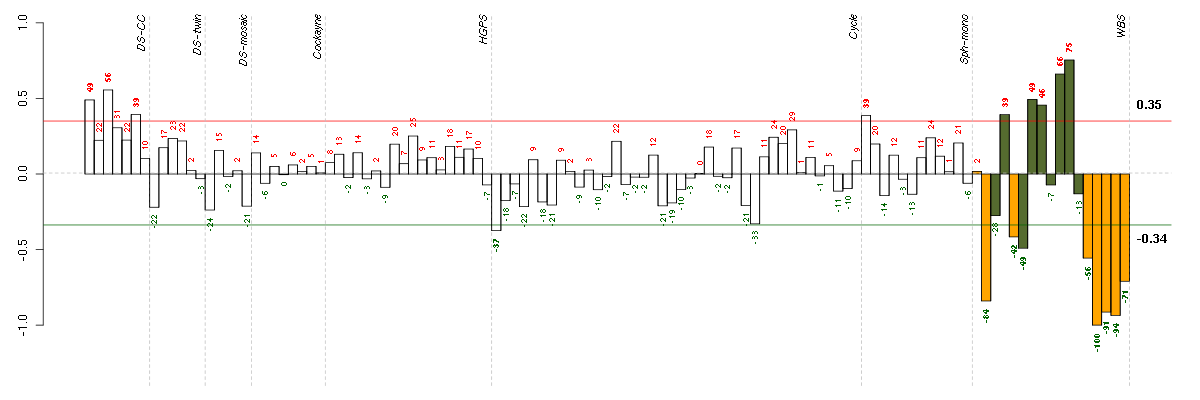
C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: 0.82 DBC1deleted in bladder cancer 1 (205818_at), score: 0.91 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.8 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.81 FLGfilaggrin (215704_at), score: 0.7 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.74 GOLSYNGolgi-localized protein (218692_at), score: 0.82 ITIH5inter-alpha (globulin) inhibitor H5 (219064_at), score: 1 NRCAMneuronal cell adhesion molecule (204105_s_at), score: 0.77 PLGLB2plasminogen-like B2 (205871_at), score: 0.72 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.72 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 5850_CNTL.CEL | 8 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |