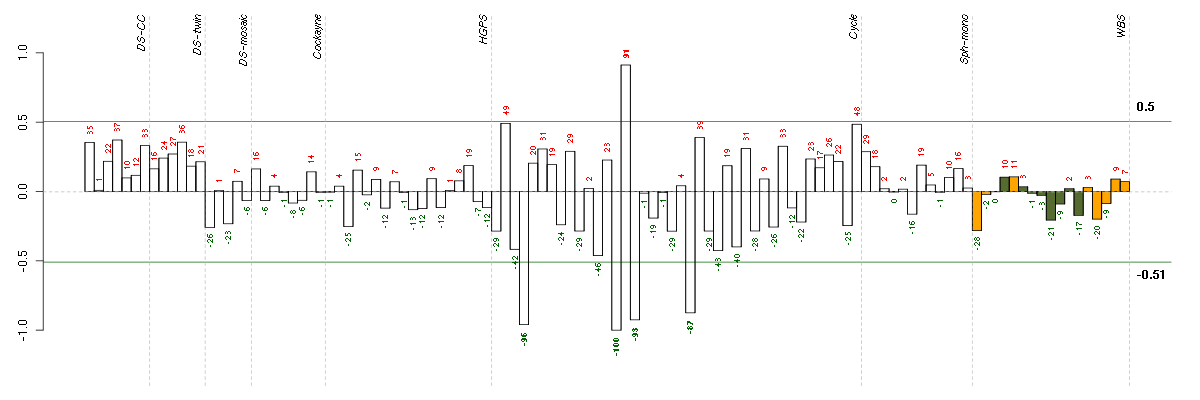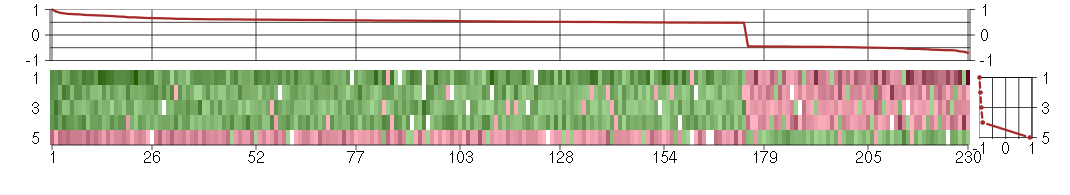

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

A1CFAPOBEC1 complementation factor (220951_s_at), score: 0.53 ACAD10acyl-Coenzyme A dehydrogenase family, member 10 (219986_s_at), score: 0.56 ACADSacyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain (202366_at), score: 0.52 ACVR1Bactivin A receptor, type IB (213198_at), score: 0.48 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.64 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.55 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.59 ADORA1adenosine A1 receptor (216220_s_at), score: 0.48 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.51 ALPLalkaline phosphatase, liver/bone/kidney (215783_s_at), score: 0.51 AMTaminomethyltransferase (204294_at), score: 0.53 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.6 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: -0.45 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: 0.59 ARSAarylsulfatase A (204443_at), score: -0.46 ASB13ankyrin repeat and SOCS box-containing 13 (218862_at), score: 0.58 ASTN2astrotactin 2 (209693_at), score: 0.51 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.54 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214244_s_at), score: 0.58 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: 0.5 BAIAP3BAI1-associated protein 3 (216356_x_at), score: 0.58 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.5 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.52 C10orf110chromosome 10 open reading frame 110 (220703_at), score: 0.5 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.48 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.47 C1orf61chromosome 1 open reading frame 61 (205103_at), score: 0.51 C1orf66chromosome 1 open reading frame 66 (218914_at), score: 0.6 C20orf12chromosome 20 open reading frame 12 (219951_s_at), score: 0.51 C2orf72chromosome 2 open reading frame 72 (213143_at), score: 0.66 C3AR1complement component 3a receptor 1 (209906_at), score: 0.49 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: 0.61 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.52 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.45 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.56 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.48 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.49 CD28CD28 molecule (206545_at), score: -0.5 CD40CD40 molecule, TNF receptor superfamily member 5 (215346_at), score: 0.59 CD53CD53 molecule (203416_at), score: 0.72 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.56 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.49 CHKAcholine kinase alpha (204233_s_at), score: 0.5 CHPcalcium binding protein P22 (214665_s_at), score: 0.48 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.59 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.59 CNNM1cyclin M1 (220166_at), score: 0.61 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.54 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.65 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.45 COX16COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) (217645_at), score: 0.5 CPA2carboxypeptidase A2 (pancreatic) (206212_at), score: 0.51 CPNE3copine III (202118_s_at), score: -0.45 CRHR1corticotropin releasing hormone receptor 1 (208593_x_at), score: 0.66 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: -0.46 CTSScathepsin S (202901_x_at), score: -0.46 CTTNcortactin (201059_at), score: 0.49 CYTH4cytohesin 4 (219183_s_at), score: 0.56 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.47 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: 0.57 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (218579_s_at), score: -0.48 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.54 DSPPdentin sialophosphoprotein (221681_s_at), score: 1 DUSP8dual specificity phosphatase 8 (206374_at), score: 0.67 EDAectodysplasin A (211130_x_at), score: 0.61 EDA2Rectodysplasin A2 receptor (221399_at), score: -0.56 ELA3Aelastase 3A, pancreatic (211738_x_at), score: 0.59 ENO3enolase 3 (beta, muscle) (204483_at), score: 0.55 EPB41L5erythrocyte membrane protein band 4.1 like 5 (220977_x_at), score: 0.61 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.6 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.56 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: 0.82 FBRSfibrosin (218255_s_at), score: 0.79 FBXO2F-box protein 2 (219305_x_at), score: 0.74 FBXO22F-box protein 22 (219638_at), score: -0.45 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.63 FGF4fibroblast growth factor 4 (206783_at), score: 0.56 FGL1fibrinogen-like 1 (205305_at), score: 0.81 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.49 FKRPfukutin related protein (219853_at), score: -0.48 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: -0.51 GANgigaxonin (220124_at), score: -0.45 GDF9growth differentiation factor 9 (221314_at), score: 0.55 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.68 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.49 GLMNglomulin, FKBP associated protein (207153_s_at), score: 0.49 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: 0.55 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: 0.62 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.59 GPR144G protein-coupled receptor 144 (216289_at), score: 0.92 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: 0.57 GSTA1glutathione S-transferase alpha 1 (215766_at), score: 0.51 HAB1B1 for mucin (215778_x_at), score: 0.81 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.66 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.69 HCG8HLA complex group 8 (215985_at), score: -0.7 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.6 HOXB5homeobox B5 (205601_s_at), score: 0.49 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.76 IGFALSinsulin-like growth factor binding protein, acid labile subunit (215712_s_at), score: 0.56 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: 0.77 IKZF4IKAROS family zinc finger 4 (Eos) (208472_at), score: 0.55 ING4inhibitor of growth family, member 4 (48825_at), score: 0.5 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.83 ITGB3integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) (204628_s_at), score: 0.54 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: 0.48 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.46 JTV1JTV1 gene (209972_s_at), score: 0.54 KIAA0892KIAA0892 (212505_s_at), score: -0.49 KIF26Bkinesin family member 26B (220002_at), score: 0.56 KLF12Kruppel-like factor 12 (208467_at), score: -0.58 KLHL4kelch-like 4 (Drosophila) (214591_at), score: -0.51 KRI1KRI1 homolog (S. cerevisiae) (218798_at), score: 0.51 KRT33Akeratin 33A (208483_x_at), score: -0.47 LAT2linker for activation of T cells family, member 2 (211768_at), score: 0.56 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.78 LMF2lipase maturation factor 2 (212682_s_at), score: -0.46 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: 0.6 LOC100133572similar to seven transmembrane helix receptor (215770_at), score: -0.46 LOC149501similar to keratin 8 (216821_at), score: 0.69 LOC391132similar to hCG2041276 (216177_at), score: 0.54 LOC80054hypothetical LOC80054 (220465_at), score: 0.59 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: 0.6 MANBAmannosidase, beta A, lysosomal (203778_at), score: 0.49 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.55 MATN3matrilin 3 (206091_at), score: -0.65 MERTKc-mer proto-oncogene tyrosine kinase (211913_s_at), score: 0.61 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: -0.61 MGC5590hypothetical protein MGC5590 (220931_at), score: -0.47 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.46 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: 0.52 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.53 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.59 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.66 NAIPNLR family, apoptosis inhibitory protein (204860_s_at), score: -0.45 NCAM2neural cell adhesion molecule 2 (205669_at), score: 0.56 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.49 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.53 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.52 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.52 NXPH3neurexophilin 3 (221991_at), score: 0.48 PAK2p21 protein (Cdc42/Rac)-activated kinase 2 (208878_s_at), score: -0.48 PANK4pantothenate kinase 4 (218771_at), score: -0.47 PCDH11Yprotocadherin 11 Y-linked (211227_s_at), score: -0.54 PCDH24protocadherin 24 (220186_s_at), score: 0.56 PDE12phosphodiesterase 12 (214826_at), score: 0.49 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.72 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: 0.62 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: 0.51 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: 0.59 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.6 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.87 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.62 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: 0.56 PRAMEF11PRAME family member 11 (217365_at), score: 0.49 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: 0.48 PRLHprolactin releasing hormone (221443_x_at), score: 0.52 PRR14proline rich 14 (218714_at), score: -0.45 PRSS3protease, serine, 3 (207463_x_at), score: -0.45 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.85 PXNpaxillin (211823_s_at), score: -0.51 R3HDM2R3H domain containing 2 (203831_at), score: 0.53 RAB30RAB30, member RAS oncogene family (206530_at), score: 0.51 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: 0.69 RBM12BRNA binding motif protein 12B (51228_at), score: -0.45 RBM38RNA binding motif protein 38 (212430_at), score: 0.63 RECQL5RecQ protein-like 5 (34063_at), score: -0.5 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: 0.6 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.55 RNF121ring finger protein 121 (219021_at), score: 0.51 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.53 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: 0.56 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.56 RPL27Aribosomal protein L27a (212044_s_at), score: 0.54 RPL38ribosomal protein L38 (202028_s_at), score: 0.6 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (220738_s_at), score: -0.59 SAA4serum amyloid A4, constitutive (207096_at), score: 0.73 SEPT5septin 5 (209767_s_at), score: 0.63 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.78 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.75 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.49 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: 0.48 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.49 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.5 SLC43A1solute carrier family 43, member 1 (204394_at), score: 0.49 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.51 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.52 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.6 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: 0.56 SLCO2A1solute carrier organic anion transporter family, member 2A1 (204368_at), score: 0.63 SPATA2Lspermatogenesis associated 2-like (214965_at), score: 0.51 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: 0.62 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.5 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.77 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.48 STIM1stromal interaction molecule 1 (202764_at), score: -0.52 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.6 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.62 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.5 TAOK2TAO kinase 2 (204986_s_at), score: -0.46 TBX21T-box 21 (220684_at), score: 0.51 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.49 TMC5transmembrane channel-like 5 (219580_s_at), score: 0.55 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.61 TMEM40transmembrane protein 40 (219503_s_at), score: 0.59 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.58 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: 0.57 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.63 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.59 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 0.57 TRAPPC9trafficking protein particle complex 9 (221836_s_at), score: 0.51 TULP2tubby like protein 2 (206733_at), score: 0.65 TWISTNBTWIST neighbor (214729_at), score: -0.58 TXNthioredoxin (216609_at), score: 0.65 VASH1vasohibin 1 (203940_s_at), score: 0.48 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.52 WDR60WD repeat domain 60 (219251_s_at), score: 0.58 WDR91WD repeat domain 91 (218971_s_at), score: 0.54 WDTC1WD and tetratricopeptide repeats 1 (215497_s_at), score: 0.57 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.55 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.58 ZNF174zinc finger protein 174 (210290_at), score: 0.62 ZNF192zinc finger protein 192 (206579_at), score: -0.46 ZNF334zinc finger protein 334 (220022_at), score: -0.6 ZNF460zinc finger protein 460 (216279_at), score: -0.51 ZNF516zinc finger protein 516 (203604_at), score: 0.52 ZNF529zinc finger protein 529 (215307_at), score: 0.52 ZNF652zinc finger protein 652 (205594_at), score: 0.53 ZNF749zinc finger protein 749 (215289_at), score: -0.45
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |