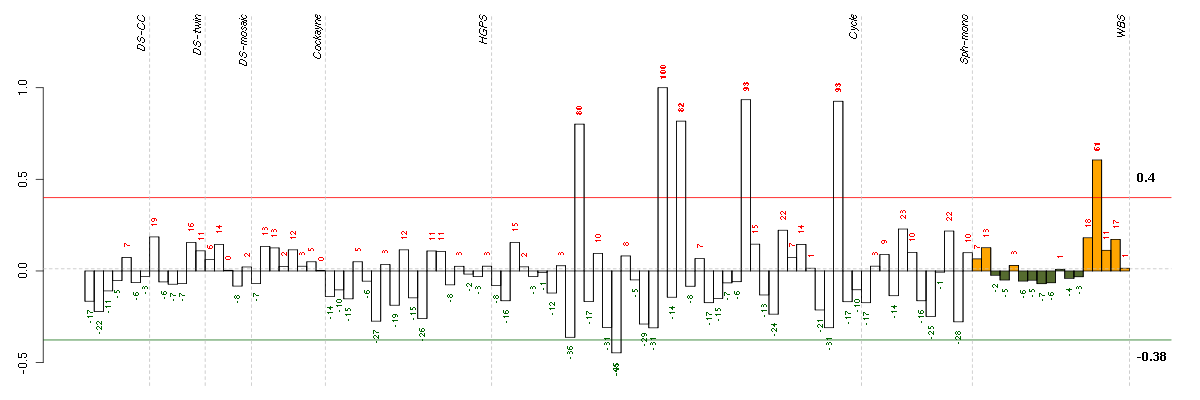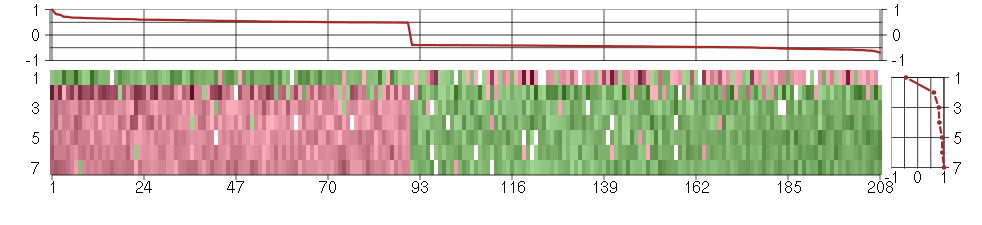

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain.
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
all
This term is the most general term possible
ACANaggrecan (205679_x_at), score: 0.68 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.53 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.41 AFF2AF4/FMR2 family, member 2 (206105_at), score: -0.46 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.5 ANXA10annexin A10 (210143_at), score: -0.57 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.49 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.57 ARHGAP8Rho GTPase activating protein 8 (37117_at), score: -0.4 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.54 ASPNasporin (219087_at), score: -0.4 ASTN2astrotactin 2 (209693_at), score: 0.68 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.5 B9D1B9 protein domain 1 (210534_s_at), score: 0.53 BAXBCL2-associated X protein (211833_s_at), score: 0.52 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.62 C11orf51chromosome 11 open reading frame 51 (204218_at), score: 0.55 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.5 C12orf5chromosome 12 open reading frame 5 (219099_at), score: 0.52 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.58 C1orf115chromosome 1 open reading frame 115 (218546_at), score: -0.46 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.51 C4orf29chromosome 4 open reading frame 29 (219980_at), score: -0.4 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.54 CAPN3calpain 3, (p94) (210944_s_at), score: -0.4 CCDC44coiled-coil domain containing 44 (221069_s_at), score: 0.51 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.48 CD320CD320 molecule (218529_at), score: 0.62 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.59 CDCP1CUB domain containing protein 1 (218451_at), score: -0.5 CDH18cadherin 18, type 2 (206280_at), score: 0.5 CEP70centrosomal protein 70kDa (219036_at), score: 0.55 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.49 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.44 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.49 CLK1CDC-like kinase 1 (214683_s_at), score: -0.45 CLK4CDC-like kinase 4 (210346_s_at), score: -0.46 CLUclusterin (208791_at), score: -0.55 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.45 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.47 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.5 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.43 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.52 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.53 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (221550_at), score: 0.54 CPA4carboxypeptidase A4 (205832_at), score: 0.59 CST6cystatin E/M (206595_at), score: -0.46 CTSFcathepsin F (203657_s_at), score: -0.43 CTSOcathepsin O (203758_at), score: -0.41 CTSScathepsin S (202901_x_at), score: -0.57 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.41 CYCSP52cytochrome c, somatic pseudogene 52 (217206_at), score: -0.42 CYorf14chromosome Y open reading frame 14 (207063_at), score: -0.44 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.55 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.42 DCPSdecapping enzyme, scavenger (218774_at), score: 0.49 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.61 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.42 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.55 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.65 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.44 DUSP6dual specificity phosphatase 6 (208892_s_at), score: -0.47 EDNRBendothelin receptor type B (204271_s_at), score: -0.43 EGFL6EGF-like-domain, multiple 6 (219454_at), score: -0.44 EMCNendomucin (219436_s_at), score: 0.52 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.56 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: -0.4 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.41 EVCEllis van Creveld syndrome (219432_at), score: -0.39 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.55 EXOSC5exosome component 5 (218481_at), score: 0.55 FAM149B1family with sequence similarity 149, member B1 (213896_x_at), score: 0.49 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: 0.53 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.43 FBXO31F-box protein 31 (219785_s_at), score: 0.58 FDXRferredoxin reductase (207813_s_at), score: 0.5 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.41 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.57 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.53 GAAglucosidase, alpha; acid (202812_at), score: -0.46 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.57 GKglycerol kinase (207387_s_at), score: -0.39 GPM6Bglycoprotein M6B (209170_s_at), score: -0.53 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.48 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.72 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: 0.51 GTF3C3general transcription factor IIIC, polypeptide 3, 102kDa (218343_s_at), score: 0.5 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.46 HOXD1homeobox D1 (205975_s_at), score: -0.39 HPCAL4hippocalcin like 4 (219671_at), score: -0.49 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (203285_s_at), score: -0.45 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.56 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: -0.45 IER2immediate early response 2 (202081_at), score: -0.4 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.59 IL33interleukin 33 (209821_at), score: -0.69 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.51 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.57 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.63 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.54 KIAA0586KIAA0586 (205631_at), score: 0.54 KIAA0907KIAA0907 (202220_at), score: -0.43 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.6 KRT17keratin 17 (212236_x_at), score: -0.4 KRT33Akeratin 33A (208483_x_at), score: -0.56 KRT9keratin 9 (208188_at), score: -0.4 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.6 LOC390940similar to R28379_1 (213556_at), score: 0.56 LOC729806similar to hCG1725380 (217544_at), score: -0.47 MAP4K4mitogen-activated protein kinase kinase kinase kinase 4 (218181_s_at), score: -0.47 MAP7D3MAP7 domain containing 3 (219576_at), score: 0.5 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.4 MLPHmelanophilin (218211_s_at), score: 0.67 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.46 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.47 MPImannose phosphate isomerase (202472_at), score: 0.6 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.57 MYO5Cmyosin VC (218966_at), score: -0.41 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: 0.6 NAG18NAG18 mRNA (210496_at), score: -0.42 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.58 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.42 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.43 NDRG4NDRG family member 4 (209159_s_at), score: 0.61 NEFLneurofilament, light polypeptide (221805_at), score: -0.56 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.47 NOL8nucleolar protein 8 (218244_at), score: 0.5 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.64 NTNG1netrin G1 (206713_at), score: -0.4 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.52 NUMBnumb homolog (Drosophila) (207545_s_at), score: -0.42 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.4 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.59 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.51 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.6 PEG3paternally expressed 3 (209242_at), score: -0.47 PENKproenkephalin (213791_at), score: -0.44 PET112LPET112-like (yeast) (204300_at), score: 0.59 PHKA1phosphorylase kinase, alpha 1 (muscle) (205450_at), score: -0.43 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: -0.41 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.48 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.56 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204286_s_at), score: -0.48 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.54 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.49 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.4 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.55 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.46 PRSS3protease, serine, 3 (207463_x_at), score: -0.61 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.57 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.49 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.59 RAB13RAB13, member RAS oncogene family (202252_at), score: 0.5 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.66 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.41 RASA2RAS p21 protein activator 2 (206636_at), score: -0.39 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.51 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: -0.55 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.4 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.6 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.43 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.64 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203788_s_at), score: -0.51 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.5 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.41 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.45 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: 0.5 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.63 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: -0.48 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.44 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: -0.41 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.4 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: -0.43 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.81 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.49 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.47 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.63 SUOXsulfite oxidase (204067_at), score: 0.68 SYNMsynemin, intermediate filament protein (212730_at), score: 0.65 SYT11synaptotagmin XI (209198_s_at), score: 0.71 TAAR2trace amine associated receptor 2 (221394_at), score: -0.44 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.52 TBCEtubulin folding cofactor E (203714_s_at), score: 0.49 TBX5T-box 5 (207155_at), score: 0.5 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.57 TGFAtransforming growth factor, alpha (205016_at), score: -0.46 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.66 TMEM158transmembrane protein 158 (213338_at), score: -0.48 TMEM160transmembrane protein 160 (219219_at), score: 0.56 TMEM22transmembrane protein 22 (219569_s_at), score: -0.41 TMEM9BTMEM9 domain family, member B (218065_s_at), score: -0.42 TMOD1tropomodulin 1 (203661_s_at), score: -0.43 TMSB15Athymosin beta 15a (205347_s_at), score: 0.8 TOMM22translocase of outer mitochondrial membrane 22 homolog (yeast) (217960_s_at), score: 0.5 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.64 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.54 TRAPPC10trafficking protein particle complex 10 (209412_at), score: -0.42 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: 0.49 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.51 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: 0.52 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.52 XPO4exportin 4 (218479_s_at), score: 0.62 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.54 ZNF219zinc finger protein 219 (219314_s_at), score: -0.54 ZNF287zinc finger protein 287 (216710_x_at), score: -0.44 ZNF688zinc finger protein 688 (213527_s_at), score: 0.5 ZNF749zinc finger protein 749 (215289_at), score: -0.47
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |