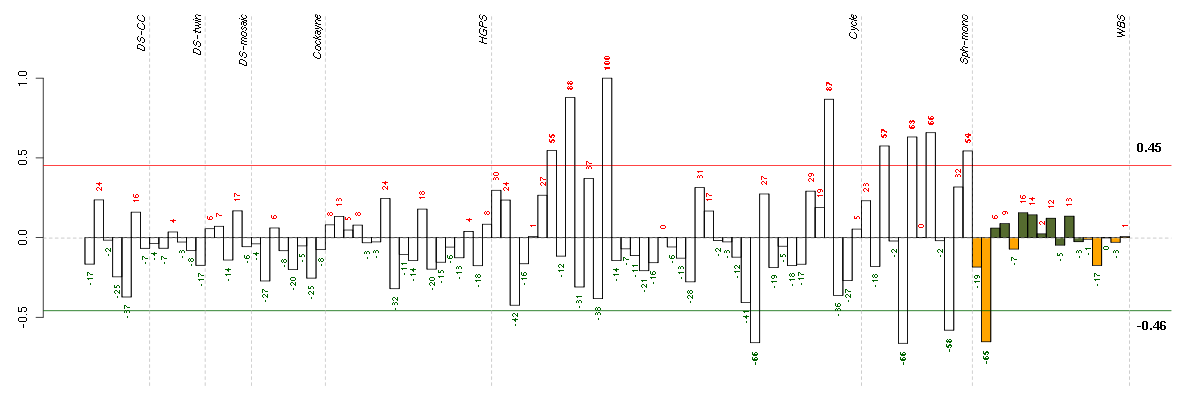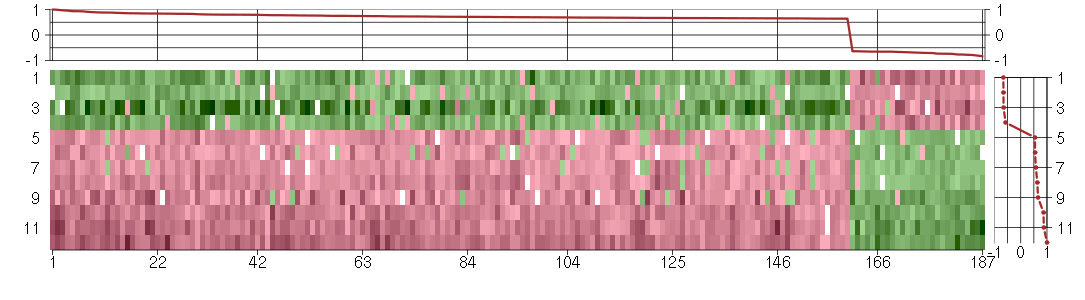

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: -0.65 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: 0.65 ABI1abl-interactor 1 (209028_s_at), score: 0.66 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.84 ADAM9ADAM metallopeptidase domain 9 (meltrin gamma) (202381_at), score: 0.66 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.71 ADORA2Badenosine A2b receptor (205891_at), score: -0.81 AHI1Abelson helper integration site 1 (221569_at), score: 1 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.73 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: -0.69 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: 0.73 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: 0.76 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (218098_at), score: 0.66 ARFIP1ADP-ribosylation factor interacting protein 1 (218230_at), score: 0.68 ATF1activating transcription factor 1 (222103_at), score: 0.78 ATMINATM interactor (201855_s_at), score: 0.69 ATP11BATPase, class VI, type 11B (212536_at), score: 0.64 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.84 AZI25-azacytidine induced 2 (218043_s_at), score: 0.76 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.75 C10orf18chromosome 10 open reading frame 18 (218331_s_at), score: 0.75 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.73 C14orf104chromosome 14 open reading frame 104 (219166_at), score: 0.68 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.72 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.72 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.73 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.85 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.86 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: 0.69 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.65 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.81 CCDC76coiled-coil domain containing 76 (219130_at), score: 0.8 CCNT2cyclin T2 (204645_at), score: 0.73 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: 0.73 CLDND1claudin domain containing 1 (208925_at), score: 0.65 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: 0.7 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: 0.74 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.65 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.93 CTAGE5CTAGE family, member 5 (215930_s_at), score: 0.65 CUL5cullin 5 (203531_at), score: 0.68 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: 0.67 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.66 DMXL1Dmx-like 1 (203791_at), score: 0.66 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.88 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.84 DOPEY1dopey family member 1 (40612_at), score: 0.8 DZIP1DAZ interacting protein 1 (204557_s_at), score: 0.71 EAF2ELL associated factor 2 (219551_at), score: 0.65 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (220342_x_at), score: 0.66 EDNRAendothelin receptor type A (204464_s_at), score: 0.75 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.79 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: 0.68 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.77 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: 0.64 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.83 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.74 EVI5ecotropic viral integration site 5 (209717_at), score: 0.71 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.69 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.7 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.69 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.7 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.89 FOSL1FOS-like antigen 1 (204420_at), score: -0.74 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.72 GHRgrowth hormone receptor (205498_at), score: 0.78 GKglycerol kinase (207387_s_at), score: 0.84 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.93 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.66 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.8 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.82 H1FXH1 histone family, member X (204805_s_at), score: -0.65 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: 0.68 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.91 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.65 HJURPHolliday junction recognition protein (218726_at), score: -0.65 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.68 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.77 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: 0.77 IMPACTImpact homolog (mouse) (218637_at), score: 0.76 INSIG2insulin induced gene 2 (209566_at), score: 0.66 ITGA1integrin, alpha 1 (214660_at), score: 0.7 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: 0.7 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.81 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.99 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.78 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.74 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.65 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.75 LUMlumican (201744_s_at), score: 0.68 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (218311_at), score: 0.65 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: -0.69 MATR3matrin 3 (200626_s_at), score: 0.68 MED23mediator complex subunit 23 (218846_at), score: 0.83 MED9mediator complex subunit 9 (218372_at), score: -0.78 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.64 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.77 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.67 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.77 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.64 MYO1Bmyosin IB (212364_at), score: 0.93 NCALDneurocalcin delta (211685_s_at), score: 0.65 NEFMneurofilament, medium polypeptide (205113_at), score: 0.66 NHLRC2NHL repeat containing 2 (219353_at), score: 0.66 NLGN4Yneuroligin 4, Y-linked (207703_at), score: 0.68 NOX4NADPH oxidase 4 (219773_at), score: 0.84 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.78 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: 0.65 OSBPL8oxysterol binding protein-like 8 (212582_at), score: 0.64 PAQR3progestin and adipoQ receptor family member III (213372_at), score: 0.73 PCDH9protocadherin 9 (219737_s_at), score: 0.74 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.88 PDCLphosducin-like (204449_at), score: 0.67 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.73 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.68 PJA2praja ring finger 2 (201133_s_at), score: 0.7 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: 0.9 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.64 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.65 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.7 PVRL3poliovirus receptor-related 3 (213325_at), score: 0.66 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.8 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.78 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: 0.73 RB1retinoblastoma 1 (203132_at), score: 0.64 RND3Rho family GTPase 3 (212724_at), score: 0.64 RNF220ring finger protein 220 (219988_s_at), score: -0.71 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.8 RPS20ribosomal protein S20 (216247_at), score: 0.66 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: 0.72 SACSspastic ataxia of Charlevoix-Saguenay (sacsin) (213262_at), score: 0.7 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: 0.8 SEC24DSEC24 family, member D (S. cerevisiae) (202375_at), score: 0.65 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.75 SHOC2soc-2 suppressor of clear homolog (C. elegans) (202777_at), score: 0.64 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.75 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.97 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.67 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.67 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.87 SMAD3SMAD family member 3 (218284_at), score: -0.64 SNNstannin (218032_at), score: -0.83 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.69 SPASTspastin (209748_at), score: 0.75 SPG20spastic paraplegia 20 (Troyer syndrome) (212526_at), score: 0.67 SPHARS-phase response (cyclin-related) (206272_at), score: 0.67 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.8 STIM1stromal interaction molecule 1 (202764_at), score: -0.7 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.84 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.7 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.78 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.67 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: 0.68 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.67 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.84 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: 0.74 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: 0.66 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: 0.69 TMEM168transmembrane protein 168 (218962_s_at), score: 0.72 TMEM2transmembrane protein 2 (218113_at), score: 0.71 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.96 TMF1TATA element modulatory factor 1 (213024_at), score: 0.88 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: -0.66 TOPORStopoisomerase I binding, arginine/serine-rich (204071_s_at), score: 0.71 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: -0.73 TRAPPC2trafficking protein particle complex 2 (219351_at), score: 0.83 TRNAU1APtRNA selenocysteine 1 associated protein 1 (218977_s_at), score: -0.7 TSNAXtranslin-associated factor X (203983_at), score: 0.64 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.76 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.71 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.67 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.71 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.85 WDR44WD repeat domain 44 (219297_at), score: 0.67 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: 0.66 YDD19YDD19 protein (37079_at), score: 0.69 YIPF4Yip1 domain family, member 4 (209551_at), score: 0.72 YTHDC2YTH domain containing 2 (213077_at), score: 0.8 YTHDF3YTH domain family, member 3 (221749_at), score: 0.66 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: 0.71 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.85 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.83 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.76 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: 0.79 ZNF146zinc finger protein 146 (200050_at), score: 0.66 ZNF268zinc finger protein 268 (209989_at), score: 0.69 ZNF518Azinc finger protein 518A (204291_at), score: 0.74
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |