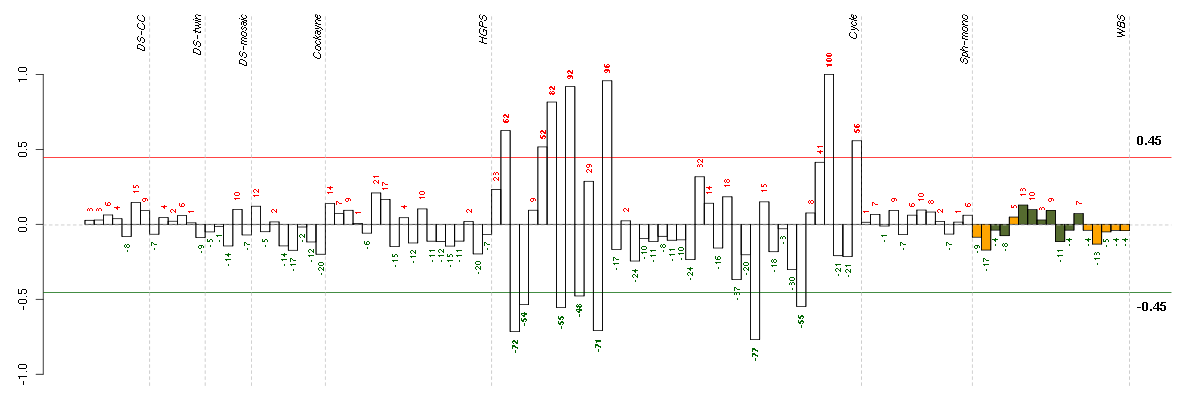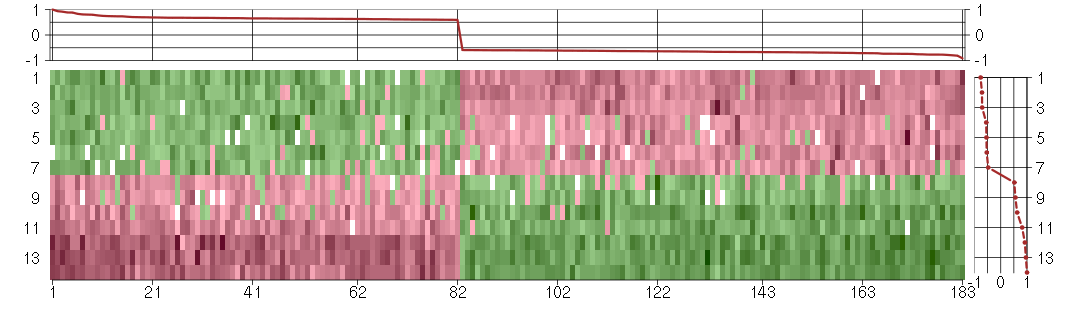

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: -0.6 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.63 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.61 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.67 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: -0.62 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.74 ADARadenosine deaminase, RNA-specific (201786_s_at), score: -0.61 ADORA2Badenosine A2b receptor (205891_at), score: -0.7 AHI1Abelson helper integration site 1 (221569_at), score: 0.61 AIM1absent in melanoma 1 (212543_at), score: -0.74 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.7 ANXA10annexin A10 (210143_at), score: 0.63 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.68 BCL2L1BCL2-like 1 (215037_s_at), score: -0.6 BMP6bone morphogenetic protein 6 (206176_at), score: 0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.62 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: 0.64 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.64 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.62 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.7 CALB2calbindin 2 (205428_s_at), score: 0.8 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.65 CAPN5calpain 5 (205166_at), score: -0.6 CASC3cancer susceptibility candidate 3 (207842_s_at), score: -0.65 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.66 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: -0.67 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.64 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: -0.78 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.6 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: 0.6 CICcapicua homolog (Drosophila) (212784_at), score: -0.66 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.71 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.6 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.59 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.7 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.77 CScitrate synthase (208660_at), score: -0.67 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: -0.6 DENND2ADENN/MADD domain containing 2A (53991_at), score: -0.6 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.6 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.65 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.68 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.88 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.68 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: -0.59 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.74 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: 0.65 EMCNendomucin (219436_s_at), score: -0.66 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.63 EPHA4EPH receptor A4 (206114_at), score: 0.68 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.66 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.77 FICDFIC domain containing (219910_at), score: 0.63 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.69 FOXK2forkhead box K2 (203064_s_at), score: -0.65 FOXM1forkhead box M1 (202580_x_at), score: -0.6 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.59 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.88 GHRgrowth hormone receptor (205498_at), score: 0.6 GKglycerol kinase (207387_s_at), score: 0.65 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.81 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.64 GMPRguanosine monophosphate reductase (204187_at), score: -0.72 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.63 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: -0.61 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.63 H1F0H1 histone family, member 0 (208886_at), score: -0.64 H1FXH1 histone family, member X (204805_s_at), score: -0.71 HAB1B1 for mucin (215778_x_at), score: 0.64 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: -0.67 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.67 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.62 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.67 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.6 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.83 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.67 IRF1interferon regulatory factor 1 (202531_at), score: -0.64 ITGA1integrin, alpha 1 (214660_at), score: 0.66 JUPjunction plakoglobin (201015_s_at), score: -0.76 KAL1Kallmann syndrome 1 sequence (205206_at), score: 1 KIAA0562KIAA0562 (204075_s_at), score: 0.61 KIAA1305KIAA1305 (220911_s_at), score: -0.69 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.68 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.62 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.6 LOC391132similar to hCG2041276 (216177_at), score: 0.65 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.65 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.61 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.61 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.7 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.8 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.71 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.68 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.68 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.67 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: 0.63 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.67 NCALDneurocalcin delta (211685_s_at), score: 0.66 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: -0.6 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.77 NEFLneurofilament, light polypeptide (221805_at), score: 0.67 NEFMneurofilament, medium polypeptide (205113_at), score: 0.93 NEO1neogenin homolog 1 (chicken) (204321_at), score: -0.64 NF1neurofibromin 1 (211094_s_at), score: -0.68 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.69 NOL10nucleolar protein 10 (218591_s_at), score: 0.61 NOX4NADPH oxidase 4 (219773_at), score: 0.67 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.6 NRXN3neurexin 3 (205795_at), score: 0.62 NXPH3neurexophilin 3 (221991_at), score: 0.6 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: -0.63 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.67 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.91 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.62 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.73 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.69 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.61 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.63 PCDH9protocadherin 9 (219737_s_at), score: 0.91 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.63 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.81 PITX1paired-like homeodomain 1 (208502_s_at), score: -0.65 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.66 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.68 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: -0.64 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.75 PTTG3pituitary tumor-transforming 3 (208511_at), score: -0.64 PXNpaxillin (211823_s_at), score: -0.62 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.77 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.62 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.73 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: -0.67 RNF220ring finger protein 220 (219988_s_at), score: -0.68 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: 0.64 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.72 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: 0.64 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.65 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: 0.64 SAP30LSAP30-like (219129_s_at), score: -0.69 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.67 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.62 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.8 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: 0.62 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.7 SEPHS1selenophosphate synthetase 1 (208939_at), score: -0.61 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.59 SF1splicing factor 1 (208313_s_at), score: -0.71 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: -0.6 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.67 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.66 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.67 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.63 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.73 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.66 SMAD3SMAD family member 3 (218284_at), score: -0.74 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.74 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.59 STIM1stromal interaction molecule 1 (202764_at), score: -0.73 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.64 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.7 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.63 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: -0.77 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.66 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.69 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: 0.63 TMEM104transmembrane protein 104 (220097_s_at), score: 0.65 TMEM2transmembrane protein 2 (218113_at), score: 0.61 TMSB15Bthymosin beta 15B (214051_at), score: -0.64 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.63 TP53tumor protein p53 (201746_at), score: -0.74 TRIM21tripartite motif-containing 21 (204804_at), score: -0.67 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.69 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: 0.61 TSKUtsukushin (218245_at), score: -0.64 ULBP2UL16 binding protein 2 (221291_at), score: 0.64 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.62 WDR4WD repeat domain 4 (221632_s_at), score: 0.63 WDR6WD repeat domain 6 (217734_s_at), score: -0.76 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.74 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: 0.6 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.66 ZNF706zinc finger protein 706 (218059_at), score: -0.6
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |