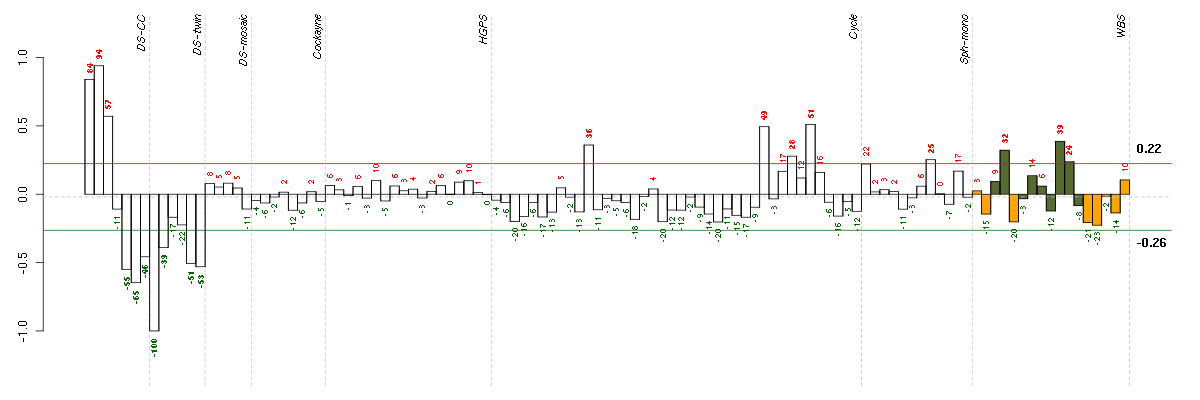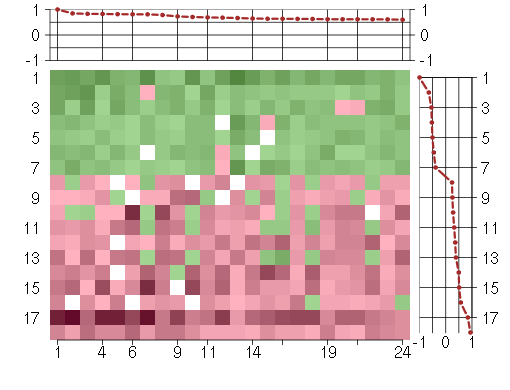

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.81 APODapolipoprotein D (201525_at), score: 0.73 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: 0.61 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.64 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: 1 DPTdermatopontin (213068_at), score: 0.63 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: 0.81 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.67 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: 0.83 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.62 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: 0.82 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: 0.65 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: 0.68 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.6 MFAP4microfibrillar-associated protein 4 (212713_at), score: 0.62 MYO1Dmyosin ID (212338_at), score: 0.62 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: 0.63 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.71 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.62 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: 0.64 PPLperiplakin (203407_at), score: 0.78 RCAN2regulator of calcineurin 2 (203498_at), score: 0.69 RRADRas-related associated with diabetes (204803_s_at), score: 0.85 WISP2WNT1 inducible signaling pathway protein 2 (205792_at), score: 0.82
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |