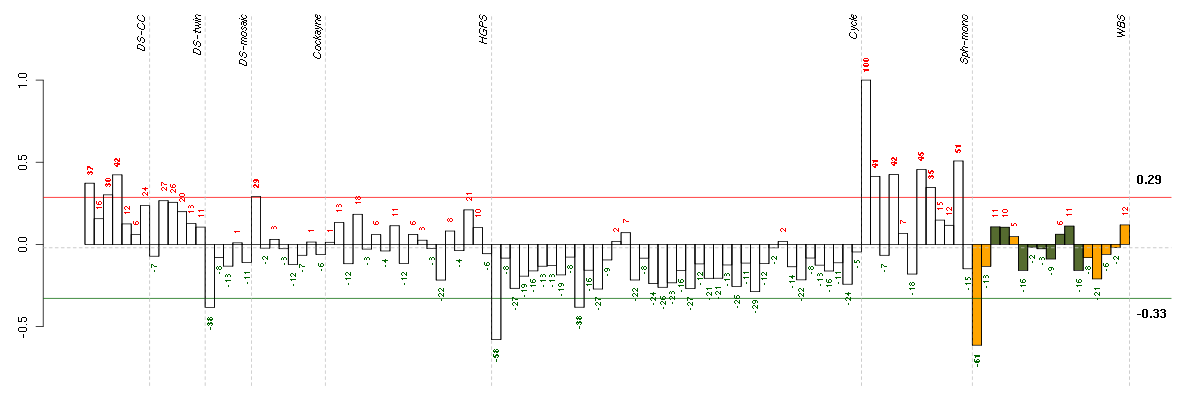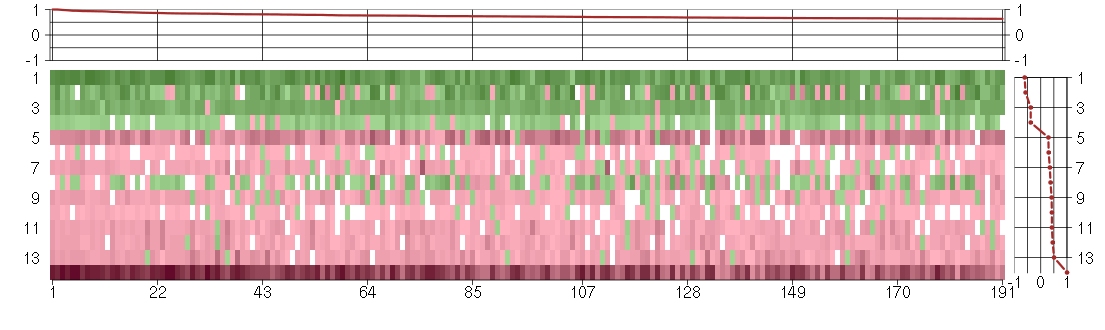

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00830 | 3.412e-03 | 0.3881 | 5 | 23 | Retinol metabolism |
| 00982 | 2.100e-02 | 0.6075 | 5 | 36 | Drug metabolism - cytochrome P450 |
| 00980 | 2.267e-02 | 0.6244 | 5 | 37 | Metabolism of xenobiotics by cytochrome P450 |
| 00591 | 2.443e-02 | 0.1519 | 3 | 9 | Linoleic acid metabolism |
ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.75 ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.89 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.66 ADAMTSL3ADAMTS-like 3 (213974_at), score: 0.65 ADORA1adenosine A1 receptor (216220_s_at), score: 0.68 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.82 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: 0.83 ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.91 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.85 ARAP2ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 (214102_at), score: 0.65 ATP5OATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (216954_x_at), score: 0.76 BCL2L11BCL2-like 11 (apoptosis facilitator) (222343_at), score: 0.64 BCL9B-cell CLL/lymphoma 9 (204129_at), score: 0.76 BTF3L2basic transcription factor 3, like 2 (217461_x_at), score: 0.64 C10orf110chromosome 10 open reading frame 110 (220703_at), score: 0.67 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.86 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.66 CADM4cell adhesion molecule 4 (222293_at), score: 0.66 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.7 CCR9chemokine (C-C motif) receptor 9 (207445_s_at), score: 0.66 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (205288_at), score: 0.68 CDR1cerebellar degeneration-related protein 1, 34kDa (207276_at), score: 0.8 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: 0.73 CEP27centrosomal protein 27kDa (220071_x_at), score: 0.74 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: 0.94 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.66 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.66 CIRBPcold inducible RNA binding protein (200811_at), score: 0.7 CLEC4EC-type lectin domain family 4, member E (219859_at), score: 0.71 COX5Bcytochrome c oxidase subunit Vb (213736_at), score: 0.72 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: 0.72 CYP1A2cytochrome P450, family 1, subfamily A, polypeptide 2 (207608_x_at), score: 0.66 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.8 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: 0.76 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.68 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.93 DAZ1deleted in azoospermia 1 (216922_x_at), score: 0.84 DAZ3deleted in azoospermia 3 (208281_x_at), score: 0.78 DAZ4deleted in azoospermia 4 (216351_x_at), score: 0.81 DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (204909_at), score: 0.69 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.72 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: 0.94 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: 0.71 DNAH7dynein, axonemal, heavy chain 7 (214222_at), score: 0.64 EEF1Deukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) (214395_x_at), score: 0.7 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.81 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: 0.7 EPAGearly lymphoid activation protein (217050_at), score: 0.73 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.79 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.72 FAM106Afamily with sequence similarity 106, member A (220575_at), score: 0.66 FAM134Afamily with sequence similarity 134, member A (222129_at), score: 0.68 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: 0.73 FBXO41F-box protein 41 (44040_at), score: 0.87 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: 0.7 FCARFc fragment of IgA, receptor for (211305_x_at), score: 0.73 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: 0.67 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 0.95 FLJ21369hypothetical protein FLJ21369 (220401_at), score: 0.72 FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.87 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: 0.76 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.63 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: 0.68 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.98 GK2glycerol kinase 2 (215430_at), score: 0.79 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.69 GRM6glutamate receptor, metabotropic 6 (208035_at), score: 0.69 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: 0.68 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.66 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.72 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.82 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.85 IFNA16interferon, alpha 16 (208448_x_at), score: 0.73 IFT20intraflagellar transport 20 homolog (Chlamydomonas) (214283_at), score: 0.64 INSRinsulin receptor (213792_s_at), score: 0.7 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.7 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.74 LAMB4laminin, beta 4 (215516_at), score: 0.7 LOC100128701heterogeneous nuclear ribonucleoprotein A1-like 2 pseudogene (216497_at), score: 0.79 LOC100128836similar to heterogeneous nuclear ribonucleoprotein A1 (217353_at), score: 0.77 LOC100129624hypothetical LOC100129624 (210748_at), score: 0.82 LOC100130233similar to NPM1 protein (216387_x_at), score: 0.8 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.66 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.76 LOC171220destrin-2 pseudogene (211325_x_at), score: 0.65 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.73 LOC51152melanoma antigen (220771_at), score: 0.65 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: 0.67 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.78 LOC728844hypothetical LOC728844 (222040_at), score: 0.77 LOC91316glucuronidase, beta/ immunoglobulin lambda-like polypeptide 1 pseudogene (215816_at), score: 0.73 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.76 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.81 MCL1myeloid cell leukemia sequence 1 (BCL2-related) (214057_at), score: 0.76 MEFVMediterranean fever (208262_x_at), score: 0.84 MEP1Bmeprin A, beta (207251_at), score: 0.68 METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.93 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.97 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.74 MKRN1makorin ring finger protein 1 (208082_x_at), score: 0.89 MLLT10myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 (216506_x_at), score: 0.71 MRPL44mitochondrial ribosomal protein L44 (218202_x_at), score: 0.74 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: 0.68 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.66 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.65 NAG18NAG18 mRNA (210496_at), score: 0.64 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.73 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.65 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.67 OR2B2olfactory receptor, family 2, subfamily B, member 2 (216408_at), score: 0.67 PBX2pre-B-cell leukemia homeobox 2 (202876_s_at), score: 0.77 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: 0.64 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.69 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.7 PER2period homolog 2 (Drosophila) (205251_at), score: 0.66 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.88 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.66 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.77 POU4F2POU class 4 homeobox 2 (207725_at), score: 0.67 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.82 PPP1R14Dprotein phosphatase 1, regulatory (inhibitor) subunit 14D (220082_at), score: 0.65 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 1 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.7 RHEBRas homolog enriched in brain (213409_s_at), score: 0.66 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: 0.64 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.71 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.7 RPL14ribosomal protein L14 (219138_at), score: 0.67 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: 0.93 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.81 RPL35Aribosomal protein L35a (215208_x_at), score: 0.85 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.73 RPS10P3ribosomal protein S10 pseudogene 3 (217336_at), score: 0.74 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: 0.77 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.81 SCD5stearoyl-CoA desaturase 5 (220232_at), score: 0.65 SCN11Asodium channel, voltage-gated, type XI, alpha subunit (220791_x_at), score: 0.76 SERF1Bsmall EDRK-rich factor 1B (centromeric) (219982_s_at), score: 0.64 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (206386_at), score: 0.7 SFNstratifin (33322_i_at), score: 0.66 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: 0.66 SFTPBsurfactant protein B (214354_x_at), score: 0.81 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.91 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: 0.74 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.63 SLC11A1solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 (217473_x_at), score: 0.72 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.67 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: 0.8 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.84 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: 0.67 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.87 SPNsialophorin (206057_x_at), score: 0.85 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 1 ST3GAL2ST3 beta-galactoside alpha-2,3-sialyltransferase 2 (217650_x_at), score: 0.64 SYCP1synaptonemal complex protein 1 (206740_x_at), score: 0.84 TAAR2trace amine associated receptor 2 (221394_at), score: 0.67 TBX6T-box 6 (207684_at), score: 0.68 TCTN2tectonic family member 2 (206438_x_at), score: 0.69 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 0.95 TNFRSF9tumor necrosis factor receptor superfamily, member 9 (207536_s_at), score: 0.67 TNPO3transportin 3 (212317_at), score: 0.73 TP53TG3TP53 target 3 (220167_s_at), score: 0.81 TP63tumor protein p63 (209863_s_at), score: 0.64 TPTEtransmembrane phosphatase with tensin homology (220205_at), score: 0.7 TRIM36tripartite motif-containing 36 (219736_at), score: 0.83 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.76 TUBA3Ctubulin, alpha 3c (210527_x_at), score: 0.7 UBQLN4ubiquilin 4 (222252_x_at), score: 0.79 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.92 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.79 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: 0.71 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.7 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: 0.75 VPRBPVpr (HIV-1) binding protein (204377_s_at), score: 0.74 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: 0.77 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.74 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.84 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.86 ZNF117zinc finger protein 117 (207605_x_at), score: 0.66 ZNF160zinc finger protein 160 (214715_x_at), score: 0.69 ZNF287zinc finger protein 287 (216710_x_at), score: 0.65 ZNF440zinc finger protein 440 (215892_at), score: 0.72 ZNF492zinc finger protein 492 (215532_x_at), score: 0.84 ZNF493zinc finger protein 493 (211064_at), score: 0.72 ZNF552zinc finger protein 552 (219741_x_at), score: 0.66 ZNF639zinc finger protein 639 (218413_s_at), score: 0.76 ZNF701zinc finger protein 701 (220242_x_at), score: 0.64 ZNF770zinc finger protein 770 (220608_s_at), score: 0.75 ZNF816Azinc finger protein 816A (217541_x_at), score: 0.89 ZPBPzona pellucida binding protein (207021_at), score: 0.65 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |