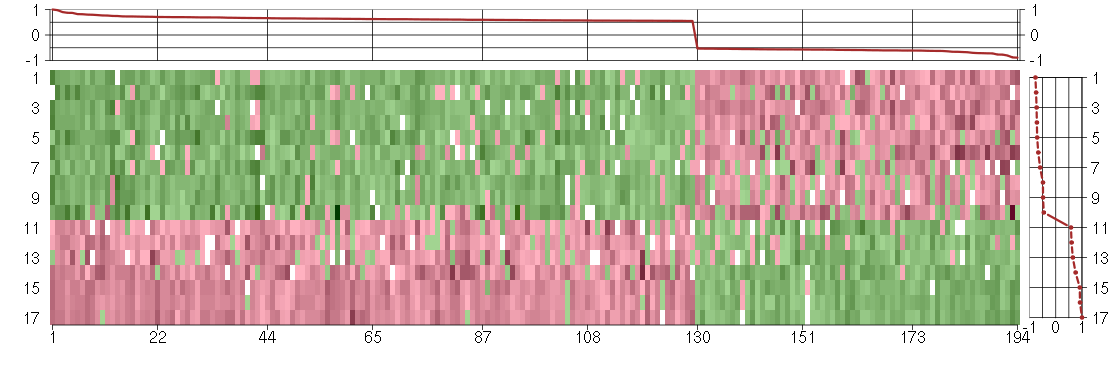

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
enzyme inhibitor activity
Stops, prevents or reduces the activity of an enzyme.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
enzyme regulator activity
Modulates the activity of an enzyme.
protease inhibitor activity
Stops, prevents or reduces the activity of a protease, any enzyme catalyzes the hydrolysis peptide bonds.
all
This term is the most general term possible
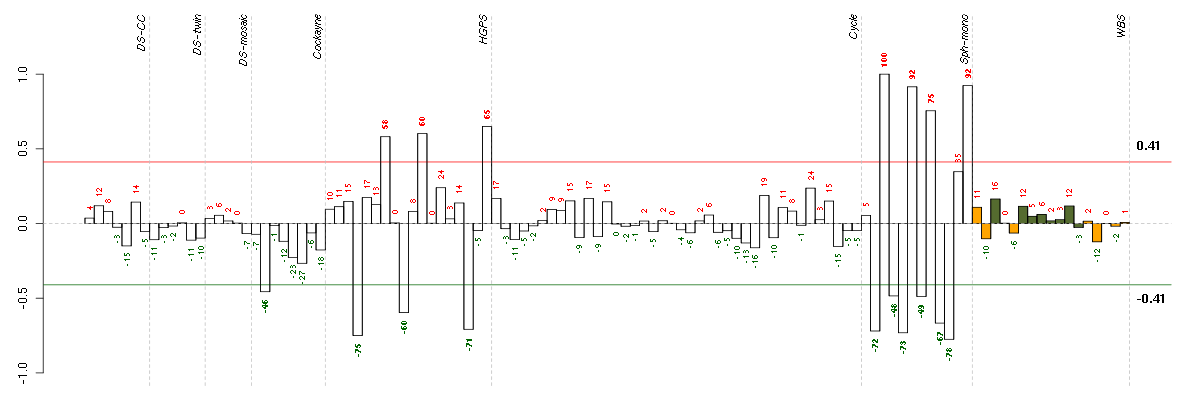
ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: -0.77 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.55 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.6 ADIPOR1adiponectin receptor 1 (217748_at), score: -0.59 ADNPactivity-dependent neuroprotector homeobox (201773_at), score: 0.57 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.68 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.67 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.71 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.57 ANPEPalanyl (membrane) aminopeptidase (202888_s_at), score: 0.62 ANXA3annexin A3 (209369_at), score: 0.59 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.6 ARHGAP29Rho GTPase activating protein 29 (203910_at), score: 0.67 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.69 ARL4CADP-ribosylation factor-like 4C (202207_at), score: -0.63 ASPNasporin (219087_at), score: 0.63 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: -0.64 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (203304_at), score: -0.6 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.63 BEX4brain expressed, X-linked 4 (215440_s_at), score: 0.61 BMP7bone morphogenetic protein 7 (209590_at), score: -0.72 BNC2basonuclin 2 (220272_at), score: 0.62 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.79 C3orf14chromosome 3 open reading frame 14 (219288_at), score: 0.65 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.62 C6orf47chromosome 6 open reading frame 47 (204968_at), score: -0.57 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.61 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.67 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.62 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.56 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.77 CEPT1choline/ethanolamine phosphotransferase 1 (219375_at), score: 0.55 CFHcomplement factor H (213800_at), score: 0.71 CFHR1complement factor H-related 1 (215388_s_at), score: 0.63 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.6 CHRDL1chordin-like 1 (209763_at), score: 0.68 CLCN4chloride channel 4 (214769_at), score: -0.54 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.66 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.66 COL3A1collagen, type III, alpha 1 (215076_s_at), score: 0.71 CRABP2cellular retinoic acid binding protein 2 (202575_at), score: 0.56 CTSDcathepsin D (200766_at), score: -0.54 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.87 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.6 DCNdecorin (211896_s_at), score: 0.61 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.55 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: 0.64 DSG2desmoglein 2 (217901_at), score: 0.56 EDEM2ER degradation enhancer, mannosidase alpha-like 2 (218282_at), score: -0.6 EDNRAendothelin receptor type A (204464_s_at), score: 0.59 EI24etoposide induced 2.4 mRNA (216396_s_at), score: -0.53 EIF4A3eukaryotic translation initiation factor 4A, isoform 3 (201303_at), score: -0.55 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.75 ENOSF1enolase superfamily member 1 (204142_at), score: 0.55 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.56 FAF1Fas (TNFRSF6) associated factor 1 (218080_x_at), score: 0.59 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.53 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.63 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.56 FBLN1fibulin 1 (202995_s_at), score: 0.72 FBXL7F-box and leucine-rich repeat protein 7 (213249_at), score: 0.58 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.66 FKBP11FK506 binding protein 11, 19 kDa (219118_at), score: 0.59 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: 0.7 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.6 FOXG1forkhead box G1 (206018_at), score: -0.56 FOXL2forkhead box L2 (220102_at), score: -0.58 FOXO1forkhead box O1 (202724_s_at), score: 0.61 FTLferritin, light polypeptide (213187_x_at), score: -0.68 FUT8fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (203988_s_at), score: 0.62 FZD6frizzled homolog 6 (Drosophila) (203987_at), score: 0.57 GALCgalactosylceramidase (204417_at), score: 0.81 GAS1growth arrest-specific 1 (204457_s_at), score: 0.58 GPR126G protein-coupled receptor 126 (213094_at), score: 0.8 GPR4G protein-coupled receptor 4 (206236_at), score: 0.59 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.56 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.72 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.61 HOXA10homeobox A10 (213150_at), score: 0.59 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: -0.57 IFI30interferon, gamma-inducible protein 30 (201422_at), score: -0.62 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (203819_s_at), score: -0.56 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.89 IL10RBinterleukin 10 receptor, beta (209575_at), score: -0.55 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.69 IRF1interferon regulatory factor 1 (202531_at), score: -0.61 ITGA8integrin, alpha 8 (214265_at), score: -0.58 JAM3junctional adhesion molecule 3 (212813_at), score: 0.74 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.62 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: 0.66 KIAA0247KIAA0247 (202181_at), score: 0.56 KIAA1797KIAA1797 (218503_at), score: 0.56 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.88 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.71 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.56 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.56 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.6 LOC654342similar to lymphocyte-specific protein 1 (214110_s_at), score: 0.56 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.87 LUMlumican (201744_s_at), score: 0.64 LXNlatexin (218729_at), score: 0.68 MAN1A1mannosidase, alpha, class 1A, member 1 (221760_at), score: 0.65 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: -0.59 MCCC1methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) (218440_at), score: -0.6 MED16mediator complex subunit 16 (43544_at), score: 0.58 MED23mediator complex subunit 23 (218846_at), score: 0.58 MED31mediator complex subunit 31 (219318_x_at), score: -0.54 MMP24matrix metallopeptidase 24 (membrane-inserted) (49679_s_at), score: -0.58 MPDZmultiple PDZ domain protein (213306_at), score: 0.58 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.7 MRPL28mitochondrial ribosomal protein L28 (204599_s_at), score: -0.57 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.64 MYH10myosin, heavy chain 10, non-muscle (212372_at), score: 0.66 MYO1Bmyosin IB (212364_at), score: 0.73 NDNnecdin homolog (mouse) (209550_at), score: 0.67 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: 0.63 NLGN1neuroligin 1 (205893_at), score: 0.7 NOX4NADPH oxidase 4 (219773_at), score: 0.77 NPTX1neuronal pentraxin I (204684_at), score: -0.61 NRN1neuritin 1 (218625_at), score: -0.62 OLIG2oligodendrocyte lineage transcription factor 2 (213825_at), score: -0.56 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: 0.7 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.57 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.6 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.59 PFTK1PFTAIRE protein kinase 1 (204604_at), score: 0.69 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.56 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.78 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.54 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: 0.7 PLCL2phospholipase C-like 2 (213309_at), score: 0.55 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.55 POPDC3popeye domain containing 3 (219926_at), score: -0.57 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.61 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.66 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.64 PTGESprostaglandin E synthase (210367_s_at), score: 0.65 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.58 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.6 RARBretinoic acid receptor, beta (205080_at), score: -0.62 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.72 RGS4regulator of G-protein signaling 4 (204337_at), score: 0.6 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: 0.58 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.66 ROBO1roundabout, axon guidance receptor, homolog 1 (Drosophila) (213194_at), score: 0.64 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: 0.62 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (212912_at), score: 0.65 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.6 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.62 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.62 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.56 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.61 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.57 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.67 SESN1sestrin 1 (218346_s_at), score: 0.56 SETD1ASET domain containing 1A (213202_at), score: -0.56 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.7 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: 0.61 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: -0.8 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: -0.57 SLC22A17solute carrier family 22, member 17 (218675_at), score: 0.69 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.57 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.64 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.64 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: -0.55 SPHARS-phase response (cyclin-related) (206272_at), score: 0.56 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: 0.62 STYK1serine/threonine/tyrosine kinase 1 (221696_s_at), score: -0.56 SULF1sulfatase 1 (212353_at), score: 0.82 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: 0.58 TCF12transcription factor 12 (208986_at), score: 0.7 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.72 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.73 THBS2thrombospondin 2 (203083_at), score: 0.9 TIMP1TIMP metallopeptidase inhibitor 1 (201666_at), score: 0.56 TMEM80transmembrane protein 80 (65630_at), score: -0.71 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.6 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.61 TPBGtrophoblast glycoprotein (203476_at), score: 0.76 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: -0.61 TRHDEthyrotropin-releasing hormone degrading enzyme (219937_at), score: 0.57 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.59 TSPAN6tetraspanin 6 (209108_at), score: 0.73 TSPYL5TSPY-like 5 (213122_at), score: 0.97 TUBB6tubulin, beta 6 (209191_at), score: 0.65 UBA52ubiquitin A-52 residue ribosomal protein fusion product 1 (221700_s_at), score: 0.57 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.54 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.69 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: -0.57 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.56 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: 0.56 ZNF365zinc finger protein 365 (206448_at), score: 0.57 ZNF518Azinc finger protein 518A (204291_at), score: 0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |